Comparative analysis of binding patterns of MADS-domain proteins in Arabidopsis thaliana
- PMID: 29940855
- PMCID: PMC6019531
- DOI: 10.1186/s12870-018-1348-8
Comparative analysis of binding patterns of MADS-domain proteins in Arabidopsis thaliana
Abstract
Background: Correct flower formation requires highly specific temporal and spatial regulation of gene expression. In Arabidopsis thaliana the majority of the master regulators that determine flower organ identity belong to the MADS-domain transcription factor family. The canonical DNA binding motif for this transcription factor family is the CArG-box, which has the consensus CC(A/T)6GG. However, so far, a comprehensive analysis of MADS-domain binding patterns has not yet been performed.
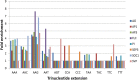
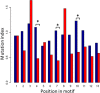
Results: Eight publicly available ChIP-seq datasets of MADS-domain proteins that regulate the floral transition and flower formation were analyzed. Surprisingly, the preferred DNA binding motif of each protein was a CArG-box with an NAA extension. Furthermore, motifs of other transcription factors were found in the vicinity of binding sites of MADS-domain transcription factors, suggesting that interaction of MADS-domain proteins with other transcription factors is important for target gene regulation. Finally, conservation of CArG-boxes between Arabidopsis ecotypes was assessed to obtain information about their evolutionary importance. CArG-boxes that fully matched the consensus were more conserved than other CArG-boxes, suggesting that the perfect CArG-box is evolutionary more important than other CArG-box variants.
Conclusion: Our analysis provides detailed insight into MADS-domain protein binding patterns. The results underline the importance of an extended version of the CArG-box and provide a first view on evolutionary conservation of MADS-domain protein binding sites in Arabidopsis ecotypes.
Keywords: CArG-box; ChIP-seq; MADS-domain proteins; Sequence conservation; Transcription factor binding specificity.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

