COP9 signalosome subunit 5A affects phenylpropanoid metabolism, trichome formation and transcription of key genes of a regulatory tri-protein complex in Arabidopsis
- PMID: 29940863
- PMCID: PMC6020244
- DOI: 10.1186/s12870-018-1347-9
COP9 signalosome subunit 5A affects phenylpropanoid metabolism, trichome formation and transcription of key genes of a regulatory tri-protein complex in Arabidopsis
Abstract
Background: Trichomes and phenylpropanoid-derived phenolics are structural and chemical protection against many adverse conditions. Their production is regulated by a network that includes a TTG1/bHLH/MYB tri-protein complex in Arabidopsis. CSN5a, encoding COP9 signalosome subunit 5a, has also been implicated in trichome and anthocyanin production; however, the regulatory roles of CSN5a in the processes through interaction with the tri-protein complex has yet to be investigated.
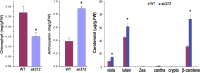
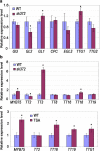
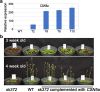
Results: In this study, a new csn5a mutant, sk372, was recovered based on its altered morphological and chemical phenotypes compared to wild-type control. Mutant characterization was conducted with an emphasis on trichome and phenylpropanoid production and possible involvement of the tri-protein complex using metabolite and gene transcription profiling and scanning electron microscopy. Seed metabolite analysis revealed that defective CSN5a led to an enhanced production of many compounds in addition to anthocyanin, most notably phenylpropanoids and carotenoids as well as a glycoside of zeatin. Consistent changes in carotenoids and anthocyanin were also found in the sk372 leaves. In addition, 370 genes were differentially expressed in 10-day old seedlings of sk372 compared to its wild type control. Real-time transcript quantitative analysis showed that in sk372, GL2 and tri-protein complex gene TT2 was significantly suppressed (p < 0.05) while complex genes EGL3 and GL3 slightly decreased (p > 0.05). Complex genes MYB75, GL1 and flavonoid biosynthetic genes TT3 and TT18 in sk372 were all significantly enhanced. Overexpression of GL3 driven by cauliflower mosaic virus 35S promotor increased the number of single pointed trichomes only, no other phenotypic recovery in sk372.
Conclusions: Our results indicated clearly that COP9 signalosome subunit CSN5a affects trichome production and the metabolism of a wide range of phenylpropanoid and carotenoid compounds. Enhanced anthocyanin accumulation and reduced trichome production were related to the enhanced MYB75 and suppressed GL2 and some other differentially expressed genes associated with the TTG1/bHLH/MYB complexes.
Keywords: Anthocyanin; Arabidopsis; COP9 signalosome; CSN5a; MYB75; Trichomes.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

