Structure and dynamics of a human myelin protein P2 portal region mutant indicate opening of the β barrel in fatty acid binding proteins
- PMID: 29940944
- PMCID: PMC6020228
- DOI: 10.1186/s12900-018-0087-2
Structure and dynamics of a human myelin protein P2 portal region mutant indicate opening of the β barrel in fatty acid binding proteins
Abstract
Background: Myelin is a multilayered proteolipid sheath wrapped around selected axons in the nervous system. Its constituent proteins play major roles in forming of the highly regular membrane structure. P2 is a myelin-specific protein of the fatty acid binding protein (FABP) superfamily, which is able to stack lipid bilayers together, and it is a target for mutations in the human inherited neuropathy Charcot-Marie-Tooth disease. A conserved residue that has been proposed to participate in membrane and fatty acid binding and conformational changes in FABPs is Phe57. This residue is thought to be a gatekeeper for the opening of the portal region upon ligand entry and egress.
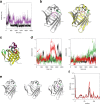
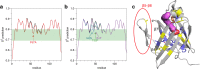
Results: We performed a structural characterization of the F57A mutant of human P2. The mutant protein was crystallized in three crystal forms, all of which showed changes in the portal region and helix α2. In addition, the behaviour of the mutant protein upon lipid bilayer binding suggested more unfolding than previously observed for wild-type P2. On the other hand, membrane binding rendered F57A heat-stable, similarly to wild-type P2. Atomistic molecular dynamics simulations showed opening of the side of the discontinuous β barrel, giving important indications on the mechanism of portal region opening and ligand entry into FABPs. The results suggest a central role for Phe57 in regulating the opening of the portal region in human P2 and other FABPs, and the F57A mutation disturbs dynamic cross-correlation networks in the portal region of P2.
Conclusions: Overall, the F57A variant presents similar properties to the P2 patient mutations recently linked to Charcot-Marie-Tooth disease. Our results identify Phe57 as a residue regulating conformational changes that may accompany membrane surface binding and ligand exchange in P2 and other FABPs.
Keywords: Crystal structure; Fatty acid-binding protein; Membrane binding; Molecular dynamics; Mutation; Myelin; Protein stability.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


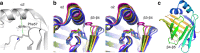

Similar articles
-
Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.J Biol Chem. 2020 Jun 26;295(26):8692-8705. doi: 10.1074/jbc.RA120.013087. Epub 2020 Apr 7. J Biol Chem. 2020. PMID: 32265298 Free PMC article.
-
Molecular mechanisms of Charcot-Marie-Tooth neuropathy linked to mutations in human myelin protein P2.Sci Rep. 2017 Jul 26;7(1):6510. doi: 10.1038/s41598-017-06781-0. Sci Rep. 2017. PMID: 28747762 Free PMC article.
-
Human myelin protein P2: from crystallography to time-lapse membrane imaging and neuropathy-associated variants.FEBS J. 2021 Dec;288(23):6716-6735. doi: 10.1111/febs.16079. Epub 2021 Jul 14. FEBS J. 2021. PMID: 34138518
-
Structural properties of the adipocyte lipid binding protein.Biochim Biophys Acta. 1999 Nov 23;1441(2-3):106-16. doi: 10.1016/s1388-1981(99)00154-7. Biochim Biophys Acta. 1999. PMID: 10570239 Review.
-
The fatty acid transport function of fatty acid-binding proteins.Biochim Biophys Acta. 2000 Jun 26;1486(1):28-44. doi: 10.1016/s1388-1981(00)00046-9. Biochim Biophys Acta. 2000. PMID: 10856711 Review.
Cited by
-
A combined computational-biophysical approach to understanding fatty acid binding to FABP7.Biophys J. 2023 Mar 7;122(5):741-752. doi: 10.1016/j.bpj.2023.02.003. Epub 2023 Feb 7. Biophys J. 2023. PMID: 36751130 Free PMC article.
-
Sub-Atomic Resolution Crystal Structures Reveal Conserved Geometric Outliers at Functional Sites.Molecules. 2019 Aug 22;24(17):3044. doi: 10.3390/molecules24173044. Molecules. 2019. PMID: 31443388 Free PMC article.
-
Ligand Entry into Fatty Acid Binding Protein via Local Unfolding Instead of Gap Widening.Biophys J. 2020 Jan 21;118(2):396-402. doi: 10.1016/j.bpj.2019.12.005. Epub 2019 Dec 14. Biophys J. 2020. PMID: 31870540 Free PMC article.
-
On the synergy between myelin proteins P0, MBP, and P2 in peripheral nerve major dense line formation.FEBS J. 2025 Aug;292(15):3960-3985. doi: 10.1111/febs.70111. Epub 2025 Apr 29. FEBS J. 2025. PMID: 40299727 Free PMC article.
-
Lipids in Pathophysiology and Development of the Membrane Lipid Therapy: New Bioactive Lipids.Membranes (Basel). 2021 Nov 24;11(12):919. doi: 10.3390/membranes11120919. Membranes (Basel). 2021. PMID: 34940418 Free PMC article. Review.
References
-
- Trapp BD, McIntyre LJ, Quarles RH, Sternberger NH, Webster HD. Immunocytochemical localization of rat peripheral nervous system myelin proteins: P2 protein is not a component of all peripheral nervous system myelin sheaths. Proc Natl Acad Sci U S A. 1979;76:3552–3556. doi: 10.1073/pnas.76.7.3552. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

