CogStack - experiences of deploying integrated information retrieval and extraction services in a large National Health Service Foundation Trust hospital
- PMID: 29941004
- PMCID: PMC6020175
- DOI: 10.1186/s12911-018-0623-9
CogStack - experiences of deploying integrated information retrieval and extraction services in a large National Health Service Foundation Trust hospital
Abstract
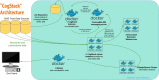
Background: Traditional health information systems are generally devised to support clinical data collection at the point of care. However, as the significance of the modern information economy expands in scope and permeates the healthcare domain, there is an increasing urgency for healthcare organisations to offer information systems that address the expectations of clinicians, researchers and the business intelligence community alike. Amongst other emergent requirements, the principal unmet need might be defined as the 3R principle (right data, right place, right time) to address deficiencies in organisational data flow while retaining the strict information governance policies that apply within the UK National Health Service (NHS). Here, we describe our work on creating and deploying a low cost structured and unstructured information retrieval and extraction architecture within King's College Hospital, the management of governance concerns and the associated use cases and cost saving opportunities that such components present.
Results: To date, our CogStack architecture has processed over 300 million lines of clinical data, making it available for internal service improvement projects at King's College London. On generated data designed to simulate real world clinical text, our de-identification algorithm achieved up to 94% precision and up to 96% recall.
Conclusion: We describe a toolkit which we feel is of huge value to the UK (and beyond) healthcare community. It is the only open source, easily deployable solution designed for the UK healthcare environment, in a landscape populated by expensive proprietary systems. Solutions such as these provide a crucial foundation for the genomic revolution in medicine.
Keywords: Clinical informatics; Elasticsearch; Electronic health records; Information extraction; Natural language processing.
Conflict of interest statement
Ethics approval and consent to participate
The creation of the CogStack software was an internal service development project for King’s College Hospital NHS Foundation Trust, and thus did not require ethical approval. As no patient identifiable data was required for the development of the software, no approval was sought from the Health Research Authority according to Confidentiality Advisory Group guidelines (
Consent for publication
Not applicable: No individual persons data is presented in this manuscript.
Competing interests
RJ and RS have received research funding from Roche, Pfizer, J&J and Lundbeck.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Simborg DW. An emerging standard for health communications: The HL7 standard. Healthc Comput Commun. 1987;4(10):58–60. - PubMed
-
- Worden R, Scott P. Simplifying HL7 Version 3 messages. Stud Health Technol Inform. 2011;169:709–13. - PubMed
-
- Antolík J. Automatic annotation of medical records. Stud Health Technol Inform. 2005;116:817–22. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

