A Deep Learning Method to Automatically Identify Reports of Scientifically Rigorous Clinical Research from the Biomedical Literature: Comparative Analytic Study
- PMID: 29941415
- PMCID: PMC6037944
- DOI: 10.2196/10281
A Deep Learning Method to Automatically Identify Reports of Scientifically Rigorous Clinical Research from the Biomedical Literature: Comparative Analytic Study
Abstract
Background: A major barrier to the practice of evidence-based medicine is efficiently finding scientifically sound studies on a given clinical topic.
Objective: To investigate a deep learning approach to retrieve scientifically sound treatment studies from the biomedical literature.
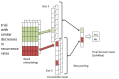
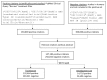
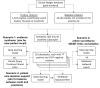
Methods: We trained a Convolutional Neural Network using a noisy dataset of 403,216 PubMed citations with title and abstract as features. The deep learning model was compared with state-of-the-art search filters, such as PubMed's Clinical Query Broad treatment filter, McMaster's textword search strategy (no Medical Subject Heading, MeSH, terms), and Clinical Query Balanced treatment filter. A previously annotated dataset (Clinical Hedges) was used as the gold standard.
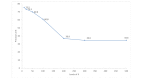
Results: The deep learning model obtained significantly lower recall than the Clinical Queries Broad treatment filter (96.9% vs 98.4%; P<.001); and equivalent recall to McMaster's textword search (96.9% vs 97.1%; P=.57) and Clinical Queries Balanced filter (96.9% vs 97.0%; P=.63). Deep learning obtained significantly higher precision than the Clinical Queries Broad filter (34.6% vs 22.4%; P<.001) and McMaster's textword search (34.6% vs 11.8%; P<.001), but was significantly lower than the Clinical Queries Balanced filter (34.6% vs 40.9%; P<.001).
Conclusions: Deep learning performed well compared to state-of-the-art search filters, especially when citations were not indexed. Unlike previous machine learning approaches, the proposed deep learning model does not require feature engineering, or time-sensitive or proprietary features, such as MeSH terms and bibliometrics. Deep learning is a promising approach to identifying reports of scientifically rigorous clinical research. Further work is needed to optimize the deep learning model and to assess generalizability to other areas, such as diagnosis, etiology, and prognosis.
Keywords: deep learning; evidence-based medicine; information retrieval; literature databases; machine learning.
©Guilherme Del Fiol, Matthew Michelson, Alfonso Iorio, Chris Cotoi, R Brian Haynes. Originally published in the Journal of Medical Internet Research (http://www.jmir.org), 25.06.2018.
Conflict of interest statement
Conflicts of Interest: MM is the Chief Scientist of InferLink Corp. and CEO of Evid Science, Inc, both of which could benefit from using the above approach as a feature within existing or new medical literature analysis products. GDF, AI, CC, and RBH have no competing interests to declare.
Figures

References
-
- Haynes RB. Where's the meat in clinical journals? ACP Journal club. 1993;119(3):A22.
-
- Ioannidis JPA. Why most published research findings are false. PLoS Med. 2005 Aug;2(8):e124. doi: 10.1371/journal.pmed.0020124. http://dx.plos.org/10.1371/journal.pmed.0020124 04-PLME-E-0321R2 - DOI - DOI - PMC - PubMed
-
- Haynes RB, Wilczynski N, McKibbon KA, Walker CJ, Sinclair JC. Developing optimal search strategies for detecting clinically sound studies in MEDLINE. J Am Med Inform Assoc. 1994;1(6):447–58. http://europepmc.org/abstract/MED/7850570 - PMC - PubMed
-
- Wilczynski NL, McKibbon KA, Haynes RB. Enhancing retrieval of best evidence for health care from bibliographic databases: calibration of the hand search of the literature. Stud Health Technol Inform. 2001;84(Pt 1):390–3. - PubMed
-
- Wilczynski NL, Morgan D, Haynes RB, Hedges Team An overview of the design and methods for retrieving high-quality studies for clinical care. BMC Med Inform Decis Mak. 2005 Jun 21;5:20. doi: 10.1186/1472-6947-5-20. https://bmcmedinformdecismak.biomedcentral.com/articles/10.1186/1472-694... 1472-6947-5-20 - DOI - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

