Stress-Induced, Highly Efficient, Donor Cell-Dependent Cell-to-Cell Natural Transformation in Bacillus subtilis
- PMID: 29941421
- PMCID: PMC6088163
- DOI: 10.1128/JB.00267-18
Stress-Induced, Highly Efficient, Donor Cell-Dependent Cell-to-Cell Natural Transformation in Bacillus subtilis
Abstract
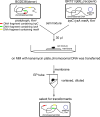
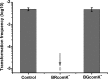
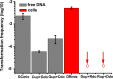
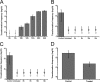
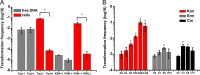
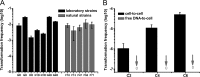
Horizontal gene transfer (HGT) is a driving force for bacterial evolution that occurs via conjugation, transduction, and transformation. Whereas conjugation and transduction depend on nonbacterial vehicles, transformation is considered a naturally occurring process in which naked DNA molecules are taken up by a competent recipient cell. Here, we report that HGT occurred between two Bacillus subtilis strains cocultured on a minimum medium agar plate for 10 h. This process was almost completely resistant to DNase treatment and appeared to require close proximity between cells. The deletion of comK in the recipient completely abolished gene transfer, indicating that the process involved transformation. This process was also highly efficient, reaching 1.75 × 106 transformants/μg DNA compared to 5.3 × 103 and 1.86 × 105 transformants/μg DNA for DNA-to-cell transformation by the same agar method and the standard two-step procedure, respectively. Interestingly, when three distantly localized chromosomal markers were selected simultaneously, the efficiency of cell-to-cell transformation still reached 6.26 × 104 transformants/μg DNA, whereas no transformants were obtained when free DNA was used as the donor. Stresses, such as starvation and exposure to antibiotics, further enhanced transformation efficiency by affecting the donor cells, suggesting that stress served as an important signal for promoting this type of HGT. Taken together, our results defined a bona fide process of cell-to-cell natural transformation (CTCNT) in B. subtilis and related species. This finding reveals the previously unrecognized role of donor cells in bacterial natural transformation and improves our understanding of how HGT drives bacterial evolution at a mechanistic level.IMPORTANCE Because DNA is easily prepared, studies of bacterial natural genetic transformation traditionally focus on recipient cells. However, such laboratory artifacts cannot explain how this process occurs in nature. In most cases, competence is only transient and involves approximately 20 to 50 genes, and it is unreasonable for bacteria to spend so many genetic resources on unpredictable and uncertain environmental DNA. Here, we characterized a donor cell-dependent CTCNT process in B. subtilis and related species that was almost completely resistant to DNase treatment and was more efficient than classical natural transformation using naked DNA as a donor, i.e., DNA-to-cell transformation, suggesting that DNA donor cells were also important in the transformation process in natural environments.
Keywords: Bacillus subtilis; cell-to-cell; donor cell; natural strains; natural transformation; stress.
Copyright © 2018 American Society for Microbiology.
Figures
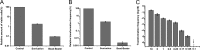
Similar articles
-
Cell-to-Cell Natural Transformation Mediated Efficient Plasmid Transfer Between Bacillus Species.Int J Mol Sci. 2025 Jan 13;26(2):621. doi: 10.3390/ijms26020621. Int J Mol Sci. 2025. PMID: 39859334 Free PMC article.
-
Cell-to-cell natural transformation in Bacillus subtilis facilitates large scale of genomic exchanges and the transfer of long continuous DNA regions.Nucleic Acids Res. 2023 May 8;51(8):3820-3835. doi: 10.1093/nar/gkad138. Nucleic Acids Res. 2023. PMID: 36912090 Free PMC article.
-
An efficient transformation method for Bacillus subtilis DB104.Appl Microbiol Biotechnol. 2012 Apr;94(2):487-93. doi: 10.1007/s00253-012-3987-2. Epub 2012 Mar 7. Appl Microbiol Biotechnol. 2012. PMID: 22395911
-
Ubiquitous late competence genes in Bacillus species indicate the presence of functional DNA uptake machineries.Environ Microbiol. 2009 Aug;11(8):1911-22. doi: 10.1111/j.1462-2920.2009.01937.x. Epub 2009 May 14. Environ Microbiol. 2009. PMID: 19453701 Review.
-
Controlling competence in Bacillus subtilis: shared use of regulators.Microbiology (Reading). 2003 Jan;149(Pt 1):9-17. doi: 10.1099/mic.0.26003-0. Microbiology (Reading). 2003. PMID: 12576575 Review.
Cited by
-
Pull in and Push Out: Mechanisms of Horizontal Gene Transfer in Bacteria.Front Microbiol. 2018 Sep 6;9:2154. doi: 10.3389/fmicb.2018.02154. eCollection 2018. Front Microbiol. 2018. PMID: 30237794 Free PMC article.
-
Cell-to-Cell Natural Transformation Mediated Efficient Plasmid Transfer Between Bacillus Species.Int J Mol Sci. 2025 Jan 13;26(2):621. doi: 10.3390/ijms26020621. Int J Mol Sci. 2025. PMID: 39859334 Free PMC article.
-
Role of natural transformation in the evolution of small cryptic plasmids in Synechocystis sp. PCC 6803.Environ Microbiol Rep. 2023 Dec;15(6):656-668. doi: 10.1111/1758-2229.13203. Epub 2023 Oct 4. Environ Microbiol Rep. 2023. PMID: 37794696 Free PMC article.
-
Cell-to-cell natural transformation in Bacillus subtilis facilitates large scale of genomic exchanges and the transfer of long continuous DNA regions.Nucleic Acids Res. 2023 May 8;51(8):3820-3835. doi: 10.1093/nar/gkad138. Nucleic Acids Res. 2023. PMID: 36912090 Free PMC article.
-
Natural Transformation of Riemerella columbina and Its Determinants.Front Microbiol. 2021 Mar 3;12:634895. doi: 10.3389/fmicb.2021.634895. eCollection 2021. Front Microbiol. 2021. PMID: 33746928 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

