Mechanism of premature translation termination on a sense codon
- PMID: 29941456
- PMCID: PMC6093235
- DOI: 10.1074/jbc.AW118.003232
Mechanism of premature translation termination on a sense codon
Abstract
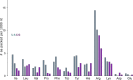
Accurate translation termination by release factors (RFs) is critical for the integrity of cellular proteomes. Premature termination on sense codons, for example, results in truncated proteins, whose accumulation could be detrimental to the cell. Nevertheless, some sense codons are prone to triggering premature termination, but the structural basis for this is unclear. To investigate premature termination, we determined a cryo-EM structure of the Escherichia coli 70S ribosome bound with RF1 in response to a UAU (Tyr) sense codon. The structure reveals that RF1 recognizes a UAU codon similarly to a UAG stop codon, suggesting that sense codons induce premature termination because they structurally mimic a stop codon. Hydrophobic interaction between the nucleobase of U3 (the third position of the UAU codon) and conserved Ile-196 in RF1 is important for misreading the UAU codon. Analyses of RNA binding in ribonucleoprotein complexes or by amino acids reveal that Ile-U packing is a frequent protein-RNA-binding motif with key functional implications. We discuss parallels with eukaryotic translation termination by the release factor eRF1.
Keywords: 70S ribosome; RNA–protein interaction; hot-spot sense codon; hydrophobic interactions; near-stop codon; ribosome function; ribosome structure; translation regulation; translation release factor; π-stacking.
© 2018 Svidritskiy et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
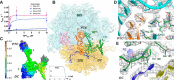

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

