O-GlcNAcylation Signal Mediates Proteasome Inhibitor Resistance in Cancer Cells by Stabilizing NRF1
- PMID: 29941490
- PMCID: PMC6094050
- DOI: 10.1128/MCB.00252-18
O-GlcNAcylation Signal Mediates Proteasome Inhibitor Resistance in Cancer Cells by Stabilizing NRF1
Abstract
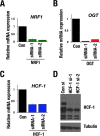
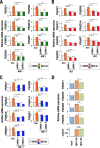
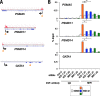
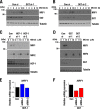
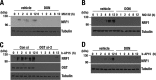
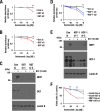
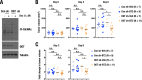
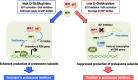
Cancer cells often heavily depend on the ubiquitin-proteasome system (UPS) for their growth and survival. Irrespective of their strong dependence on the proteasome activity, cancer cells, except for multiple myeloma, are mostly resistant to proteasome inhibitors. A major cause of this resistance is the proteasome bounce-back response mediated by NRF1, a transcription factor that coordinately activates proteasome subunit genes. To identify new targets for efficient suppression of UPS, we explored, using immunoprecipitation and mass spectrometry, the possible existence of nuclear proteins that cooperate with NRF1 and identified O-linked N-acetylglucosamine transferase (OGT) and host cell factor C1 (HCF-1) as two proteins capable of forming a complex with NRF1. O-GlcNAcylation catalyzed by OGT was essential for NRF1 stabilization and consequent upregulation of proteasome subunit genes. Meta-analysis of breast and colorectal cancers revealed positive correlations in the relative protein abundance of OGT and proteasome subunits. OGT inhibition was effective at sensitizing cancer cells to a proteasome inhibitor both in culture cells and a xenograft mouse model. Since active O-GlcNAcylation is a feature of cancer metabolism, our study has clarified a novel linkage between cancer metabolism and UPS function and added a new regulatory axis to the regulation of the proteasome activity.
Keywords: NRF1; O-GlcNAcylation; OGT; proteasome.
Copyright © 2018 Sekine et al.
Figures




Similar articles
-
Transcription factor Nrf1 is negatively regulated by its O-GlcNAcylation status.FEBS Lett. 2015 Aug 19;589(18):2347-58. doi: 10.1016/j.febslet.2015.07.030. Epub 2015 Jul 29. FEBS Lett. 2015. PMID: 26231763
-
Nuclear factor-erythroid-2 related transcription factor-1 (Nrf1) is regulated by O-GlcNAc transferase.Free Radic Biol Med. 2017 Sep;110:196-205. doi: 10.1016/j.freeradbiomed.2017.06.008. Epub 2017 Jun 15. Free Radic Biol Med. 2017. PMID: 28625484
-
HCF-1 as a key modulator of OGT function and O-GlcNAcylation in the liver.Sci Rep. 2025 Aug 3;15(1):28328. doi: 10.1038/s41598-025-11813-1. Sci Rep. 2025. PMID: 40754593 Free PMC article.
-
The Biochemistry of O-GlcNAc Transferase: Which Functions Make It Essential in Mammalian Cells?Annu Rev Biochem. 2016 Jun 2;85:631-57. doi: 10.1146/annurev-biochem-060713-035344. Annu Rev Biochem. 2016. PMID: 27294441 Review.
-
Essential role of O-GlcNAcylation in stabilization of oncogenic factors.Biochim Biophys Acta Gen Subj. 2019 Aug;1863(8):1302-1317. doi: 10.1016/j.bbagen.2019.04.002. Epub 2019 Apr 26. Biochim Biophys Acta Gen Subj. 2019. PMID: 31034911 Review.
Cited by
-
Understanding the Transcription Factor NFE2L1/NRF1 from the Perspective of Hallmarks of Cancer.Antioxidants (Basel). 2024 Jun 22;13(7):758. doi: 10.3390/antiox13070758. Antioxidants (Basel). 2024. PMID: 39061827 Free PMC article. Review.
-
DNA methylation reprograms cardiac metabolic gene expression in end-stage human heart failure.Am J Physiol Heart Circ Physiol. 2019 Oct 1;317(4):H674-H684. doi: 10.1152/ajpheart.00016.2019. Epub 2019 Jul 12. Am J Physiol Heart Circ Physiol. 2019. PMID: 31298559 Free PMC article.
-
Regulation and Functions of the ER-Associated Nrf1 Transcription Factor.Cold Spring Harb Perspect Biol. 2023 Jan 3;15(1):a041266. doi: 10.1101/cshperspect.a041266. Cold Spring Harb Perspect Biol. 2023. PMID: 35940907 Free PMC article. Review.
-
Targeting O-GlcNAcylation to overcome resistance to anti-cancer therapies.Front Oncol. 2022 Aug 17;12:960312. doi: 10.3389/fonc.2022.960312. eCollection 2022. Front Oncol. 2022. PMID: 36059648 Free PMC article. Review.
-
Functional Analysis of O-GlcNAcylation in Cancer Metastasis.Front Oncol. 2020 Oct 27;10:585288. doi: 10.3389/fonc.2020.585288. eCollection 2020. Front Oncol. 2020. PMID: 33194731 Free PMC article. Review.
References
-
- Nijhawan D, Zack TI, Ren Y, Strickland MR, Lamothe R, Schumacher SE, Tsherniak A, Besche HC, Rosenbluh J, Shehata S, Cowley GS, Weir BA, Goldberg AL, Mesirov JP, Root DE, Bhatia SN, Beroukhim R, Hahn WC. 2012. Cancer vulnerabilities unveiled by genomic loss. Cell 150:842–854. doi:10.1016/j.cell.2012.07.023. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
