Validating a 14-Drug Microtiter Plate Containing Bedaquiline and Delamanid for Large-Scale Research Susceptibility Testing of Mycobacterium tuberculosis
- PMID: 29941636
- PMCID: PMC6125532
- DOI: 10.1128/AAC.00344-18
Validating a 14-Drug Microtiter Plate Containing Bedaquiline and Delamanid for Large-Scale Research Susceptibility Testing of Mycobacterium tuberculosis
Abstract
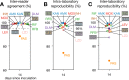
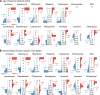
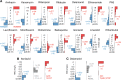
The UKMYC5 plate is a 96-well microtiter plate designed by the CRyPTIC Consortium (Comprehensive Resistance Prediction for Tuberculosis: an International Consortium) to enable the measurement of MICs of 14 different antituberculosis (anti-TB) compounds for >30,000 clinical Mycobacterium tuberculosis isolates. Unlike the MYCOTB plate, on which the UKMYC5 plate is based, the UKMYC5 plate includes two new (bedaquiline and delamanid) and two repurposed (clofazimine and linezolid) compounds. UKMYC5 plates were tested by seven laboratories on four continents by use of a panel of 19 external quality assessment (EQA) strains, including H37Rv. To assess the optimal combination of reading method and incubation time, MICs were measured from each plate by two readers, using three methods (mirrored box, microscope, and Vizion digital viewing system), after 7, 10, 14, and 21 days of incubation. In addition, all EQA strains were subjected to whole-genome sequencing and phenotypically characterized by the 7H10/7H11 agar proportion method (APM) and by use of MGIT960 mycobacterial growth indicator tubes. We concluded that the UKMYC5 plate is optimally read using the Vizion system after 14 days of incubation, achieving an interreader agreement of 97.9% and intra- and interlaboratory reproducibility rates of 95.6% and 93.1%, respectively. The mirrored box had a similar reproducibility. Strains classified as resistant by APM, MGIT960, or the presence of mutations known to confer resistance consistently showed elevated MICs compared to those for strains classified as susceptible. Finally, the UKMYC5 plate records intermediate MICs for one strain for which the APM measured MICs close to the applied critical concentration, providing early evidence that the UKMYC5 plate can quantitatively measure the magnitude of resistance to anti-TB compounds that is due to specific genetic variation.
Keywords: Mycobacterium tuberculosis; antibiotic resistance; antimicrobial agents; diagnostics.
Copyright © 2018 Rancoita et al.
Figures



References
-
- World Health Organization. 2017. Global tuberculosis report. WHO, Geneva, Switzerland.
-
- World Health Organization. 2015. The End TB strategy. WHO, Geneva, Switzerland.
-
- Franzblau SG, Witzig RS, McLaughlin JC, Torres P, Madico G, Hernandez A, Degnan MT, Cook MB, Quenzer VK, Ferguson RM, Gilman RH. 1998. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J Clin Microbiol 36:362–366. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

