A Novel Acquired t(2;4)(q36.1;q24) with a Concurrent Submicroscopic del(4)(q23q24) in An Adult with Polycythemia Vera
- PMID: 29941837
- PMCID: PMC6071118
- DOI: 10.3390/cancers10070214
A Novel Acquired t(2;4)(q36.1;q24) with a Concurrent Submicroscopic del(4)(q23q24) in An Adult with Polycythemia Vera
Abstract
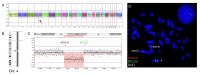
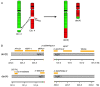
Background: Polycythemia vera (PV) is a clonal myeloid stem cell disease characterized by a growth-factor independent erythroid proliferation with an inherent tendency to transform into overt acute myeloid malignancy. Approximately 95% of the PV patients harbor the JAK2V617F mutation while less than 35% of the patients harbor cytogenetic abnormalities at the time of diagnosis. Methods and Results: Here we present a JAK2V617F positive PV patient where G-banding revealed an apparently balanced t(2;4)(q35;q21), which was confirmed by 24-color karyotyping. Oligonucleotide array-based Comparative Genomic Hybridization (aCGH) analysis revealed an interstitial 5.4 Mb large deletion at 4q23q24. Locus-specific fluorescent in situ hybridization (FISH) analyses confirmed the mono-allelic 4q deletion and that it was located on der(4)t(2;4). Additional locus-specific bacterial artificial chromosome (BAC) probes and mBanding refined the breakpoint on chromosome 2. With these methods the karyotype was revised to 46,XX,t(2;4)(q36.1;q24)[18]/46,XX[7]. Conclusions: This is the first report on a PV patient associated with an acquired novel t(2;4)(q36.1;q24) and a concurrent submicroscopic deletion del(4)(q23q24). The study also underscores the benefit of combined usage of FISH and oligo-based aCGH analysis in characterizing chromosomal abnormalities. The present findings provide additional clues to unravel important molecular pathways in PV to obtain the full spectrum of acquired chromosomal and genomic aberrations, which eventually may improve treatment options.
Keywords: chromosomal abnormality; oligonucleotide array-based Comparative Genomic Hybridization (aCGH); polycythemia vera; submicroscopic deletion del(4q).
Conflict of interest statement
The author declares no conflicts of interest.
Figures


Similar articles
-
A novel acquired cryptic three-way translocation t(2;11;5)(p21.3;q13.5;q23.2) with a submicroscopic deletion at 11p14.3 in an adult with hypereosinophilic syndrome.Exp Mol Pathol. 2015 Aug;99(1):50-5. doi: 10.1016/j.yexmp.2015.05.005. Epub 2015 May 8. Exp Mol Pathol. 2015. PMID: 25962659
-
A novel insertion ins(18;5)(q21.1;q31.2q35.1) in acute myeloid leukemia associated with microdeletions at 5q31.2, 5q35.1q35.2 and 18q12.3q21.1 detected by oligobased array comparative genomic hybridization.Mol Cytogenet. 2014 Sep 25;7(1):63. doi: 10.1186/s13039-014-0063-x. eCollection 2014. Mol Cytogenet. 2014. PMID: 25279000 Free PMC article.
-
Oligo-based aCGH analysis reveals cryptic unbalanced der(6)t(X;6) in pediatric t(12;21)-positive acute lymphoblastic leukemia.Exp Mol Pathol. 2016 Aug;101(1):38-43. doi: 10.1016/j.yexmp.2016.05.010. Epub 2016 May 20. Exp Mol Pathol. 2016. PMID: 27215399
-
Conventional cytogenetics in myelofibrosis: literature review and discussion.Eur J Haematol. 2009 May;82(5):329-38. doi: 10.1111/j.1600-0609.2009.01224.x. Epub 2009 Jan 9. Eur J Haematol. 2009. PMID: 19141119 Review.
-
Chromosome 17p13.3 deletion syndrome: aCGH characterization, prenatal findings and diagnosis, and literature review.Gene. 2013 Dec 10;532(1):152-9. doi: 10.1016/j.gene.2013.09.044. Epub 2013 Sep 19. Gene. 2013. PMID: 24055730 Review.
References
-
- The International Agency for Research on Cancer . In: WHO Classification of Tumours of Haematopoietic and Lymphoid Tissue. 4th ed. Swerdlow S., Campo E., Harris N.L., Jaffe E.S., Pileri S.A., Stein H., Thiele J., Vardiman J.W., editors. IARC Publications; Lyon, France: 2008.
-
- Gangat N., Strand J., Lasho T.L., Finke C.M., Knudson R.A., Pardanani A., Li C.Y., Ketterling R.P., Tefferi A. Cytogenetic studies at diagnosis in polycythemia vera: Clinical and JAK2V617F allele burden correlates. Eur. J. Haematol. 2008;80:197–200. doi: 10.1111/j.1600-0609.2007.01003.x. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

