Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil
- PMID: 29941893
- PMCID: PMC6018108
- DOI: 10.1038/s41598-018-27890-4
Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil
Abstract
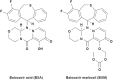
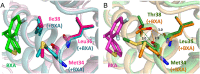
Baloxavir acid (BXA), derived from the prodrug baloxavir marboxil (BXM), potently and selectively inhibits the cap-dependent endonuclease within the polymerase PA subunit of influenza A and B viruses. In clinical trials, single doses of BXM profoundly decrease viral titers as well as alleviating influenza symptoms. Here, we characterize the impact on BXA susceptibility and replicative capacity of variant viruses detected in the post-treatment monitoring of the clinical studies. We find that the PA I38T substitution is a major pathway for reduced susceptibility to BXA, with 30- to 50-fold and 7-fold EC50 changes in A and B viruses, respectively. The viruses harboring the I38T substitution show severely impaired replicative fitness in cells, and correspondingly reduced endonuclease activity in vitro. Co-crystal structures of wild-type and I38T influenza A and B endonucleases bound to BXA show that the mutation reduces van der Waals contacts with the inhibitor. A reduced affinity to the I38T mutant is supported by the lower stability of the BXA-bound endonuclease. These mechanistic insights provide markers for future surveillance of treated populations.
Conflict of interest statement
All authors, except V.S. and S.C., were employees of Shionogi & Co, Ltd. in Osaka, Japan. The results in the paper were partially generated by the funding from Shionogi & Co., Ltd.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

