Site-specific characterization of endogenous SUMOylation across species and organs
- PMID: 29942033
- PMCID: PMC6018634
- DOI: 10.1038/s41467-018-04957-4
Site-specific characterization of endogenous SUMOylation across species and organs
Abstract
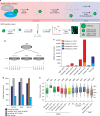
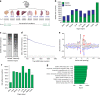
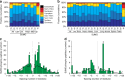
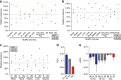
Small ubiquitin-like modifiers (SUMOs) are post-translational modifications that play crucial roles in most cellular processes. While methods exist to study exogenous SUMOylation, large-scale characterization of endogenous SUMO2/3 has remained technically daunting. Here, we describe a proteomics approach facilitating system-wide and in vivo identification of lysines modified by endogenous and native SUMO2. Using a peptide-level immunoprecipitation enrichment strategy, we identify 14,869 endogenous SUMO2/3 sites in human cells during heat stress and proteasomal inhibition, and quantitatively map 1963 SUMO sites across eight mouse tissues. Characterization of the SUMO equilibrium highlights striking differences in SUMO metabolism between cultured cancer cells and normal tissues. Targeting preferences of SUMO2/3 vary across different organ types, coinciding with markedly differential SUMOylation states of all enzymes involved in the SUMO conjugation cascade. Collectively, our systemic investigation details the SUMOylation architecture across species and organs and provides a resource of endogenous SUMOylation sites on factors important in organ-specific functions.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

