Leveraging molecular quantitative trait loci to understand the genetic architecture of diseases and complex traits
- PMID: 29942083
- PMCID: PMC6030458
- DOI: 10.1038/s41588-018-0148-2
Leveraging molecular quantitative trait loci to understand the genetic architecture of diseases and complex traits
Abstract
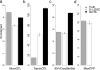
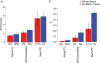
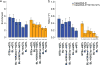
There is increasing evidence that many risk loci found using genome-wide association studies are molecular quantitative trait loci (QTLs). Here we introduce a new set of functional annotations based on causal posterior probabilities of fine-mapped molecular cis-QTLs, using data from the Genotype-Tissue Expression (GTEx) and BLUEPRINT consortia. We show that these annotations are more strongly enriched for heritability (5.84× for eQTLs; P = 1.19 × 10-31) across 41 diseases and complex traits than annotations containing all significant molecular QTLs (1.80× for expression (e)QTLs). eQTL annotations obtained by meta-analyzing all GTEx tissues generally performed best, whereas tissue-specific eQTL annotations produced stronger enrichments for blood- and brain-related diseases and traits. eQTL annotations restricted to loss-of-function intolerant genes were even more enriched for heritability (17.06×; P = 1.20 × 10-35). All molecular QTLs except splicing QTLs remained significantly enriched in joint analysis, indicating that each of these annotations is uniquely informative for disease and complex trait architectures.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- 10 Years of GWAS Discovery: Biology, Function, and Translation. - PubMed - NCBI. Available at: https://www.ncbi.nlm.nih.gov/pubmed/28686856. (Accessed: 24th March 2018) - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

