A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments
- PMID: 29942093
- PMCID: PMC6475503
- DOI: 10.1038/s41591-018-0085-8
A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments
Abstract
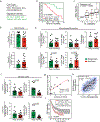
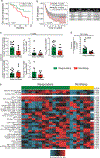
Intratumoral stimulatory dendritic cells (SDCs) play an important role in stimulating cytotoxic T cells and driving immune responses against cancer. Understanding the mechanisms that regulate their abundance in the tumor microenvironment (TME) could unveil new therapeutic opportunities. We find that in human melanoma, SDC abundance is associated with intratumoral expression of the gene encoding the cytokine FLT3LG. FLT3LG is predominantly produced by lymphocytes, notably natural killer (NK) cells in mouse and human tumors. NK cells stably form conjugates with SDCs in the mouse TME, and genetic and cellular ablation of NK cells in mice demonstrates their importance in positively regulating SDC abundance in tumor through production of FLT3L. Although anti-PD-1 'checkpoint' immunotherapy for cancer largely targets T cells, we find that NK cell frequency correlates with protective SDCs in human cancers, with patient responsiveness to anti-PD-1 immunotherapy, and with increased overall survival. Our studies reveal that innate immune SDCs and NK cells cluster together as an excellent prognostic tool for T cell-directed immunotherapy and that these innate cells are necessary for enhanced T cell tumor responses, suggesting this axis as a target for new therapies.
Figures




Comment in
-
Sequencing cells of the immune TME.Nat Rev Clin Oncol. 2018 Sep;15(9):531. doi: 10.1038/s41571-018-0069-0. Nat Rev Clin Oncol. 2018. PMID: 29985469 No abstract available.
References
Methods only References
-
- Benjamini Y & Hochberg Y Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B 57, 289–300 (1995).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

