Nucleolin modulates compartmentalization and dynamics of histone 2B-ECFP in the nucleolus
- PMID: 29943658
- PMCID: PMC6165600
- DOI: 10.1080/19491034.2018.1471936
Nucleolin modulates compartmentalization and dynamics of histone 2B-ECFP in the nucleolus
Abstract
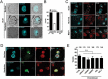
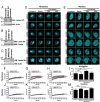
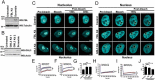
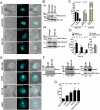
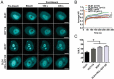
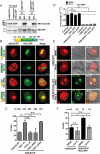
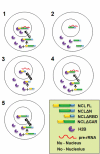
Eukaryotic cells have 2 to 3 discrete nucleoli required for ribosome synthesis. Nucleoli are phase separated nuclear sub-organelles. Here we examined the role of nuclear Lamins and nucleolar factors in modulating the compartmentalization and dynamics of histone 2B (H2B-ECFP) in the nucleolus. Live imaging and Fluorescence Recovery After Photobleaching (FRAP) of labelled H2B, showed that the depletion of Lamin B1, Fibrillarin (FBL) or Nucleostemin (GNL3), enhances H2B-ECFP mobility in the nucleolus. Furthermore, Nucleolin knockdown significantly decreases H2B-ECFP compartmentalization in the nucleolus, while H2B-ECFP residence and mobility in the nucleolus was prolonged upon Nucleolin overexpression. Co-expression of N-terminal and RNA binding domain (RBD) deletion mutants of Nucleolin or inhibiting 45S rRNA synthesis reduces the sequestration of H2B-ECFP in the nucleolus. Taken together, these studies reveal a crucial role of Nucleolin-rRNA complex in modulating the compartmentalization, stability and dynamics of H2B within the nucleolus.
Keywords: H2B; Nucleolus; lamin; nucleolin; nucleus; rRNA.
Figures
Similar articles
-
Lamin B2 Modulates Nucleolar Morphology, Dynamics, and Function.Mol Cell Biol. 2017 Nov 28;37(24):e00274-17. doi: 10.1128/MCB.00274-17. Print 2017 Dec 15. Mol Cell Biol. 2017. PMID: 28993479 Free PMC article.
-
Nucleolin functions in nucleolus formation and chromosome congression.J Cell Sci. 2007 Jun 15;120(Pt 12):2091-105. doi: 10.1242/jcs.008771. Epub 2007 May 29. J Cell Sci. 2007. PMID: 17535846
-
Nucleolar localization of aprataxin is dependent on interaction with nucleolin and on active ribosomal DNA transcription.Hum Mol Genet. 2006 Jul 15;15(14):2239-49. doi: 10.1093/hmg/ddl149. Epub 2006 Jun 15. Hum Mol Genet. 2006. PMID: 16777843
-
Plant nucleolar DNA: Green light shed on the role of Nucleolin in genome organization.Nucleus. 2017 Jan 2;8(1):11-16. doi: 10.1080/19491034.2016.1236167. Epub 2016 Sep 20. Nucleus. 2017. PMID: 27644794 Free PMC article. Review.
-
Structure and functions of nucleolin.J Cell Sci. 1999 Mar;112 ( Pt 6):761-72. doi: 10.1242/jcs.112.6.761. J Cell Sci. 1999. PMID: 10036227 Review.
Cited by
-
A reciprocal translocation involving Aspergillus nidulans snxAHrb1/Gbp2 and gyfA uncovers a new regulator of the G2-M transition and reveals a role in transcriptional repression for the setBSet2 histone H3-lysine-36 methyltransferase.Genetics. 2022 Sep 30;222(2):iyac130. doi: 10.1093/genetics/iyac130. Genetics. 2022. PMID: 36005881 Free PMC article.
-
The MYC/TXNIP axis mediates NCL-Suppressed CD8+T cell immune response in lung adenocarcinoma.Mol Med. 2025 May 9;31(1):180. doi: 10.1186/s10020-025-01224-3. Mol Med. 2025. PMID: 40346484 Free PMC article.
-
Antagonising Chromatin Remodelling Activities in the Regulation of Mammalian Ribosomal Transcription.Genes (Basel). 2021 Jun 24;12(7):961. doi: 10.3390/genes12070961. Genes (Basel). 2021. PMID: 34202617 Free PMC article. Review.
-
A novel role for nucleolin in splice site selection.RNA Biol. 2022;19(1):333-352. doi: 10.1080/15476286.2021.2020455. Epub 2021 Dec 31. RNA Biol. 2022. PMID: 35220879 Free PMC article.
-
Plant Histone HTB (H2B) Variants in Regulating Chromatin Structure and Function.Plants (Basel). 2020 Oct 25;9(11):1435. doi: 10.3390/plants9111435. Plants (Basel). 2020. PMID: 33113795 Free PMC article. Review.
References
-
- Phair RD, Misteli T.. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
