A palmitoyltransferase Approximated gene Bm-app regulates wing development in Bombyx mori
- PMID: 29943911
- PMCID: PMC7379679
- DOI: 10.1111/1744-7917.12629
A palmitoyltransferase Approximated gene Bm-app regulates wing development in Bombyx mori
Abstract
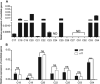
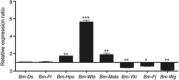
The silkworm Bombyx mori is an important lepidopteran model insect in which many kinds of natural mutants have been identified. However, molecular mechanisms of most of these mutants remain to be explored. Here we report the identification of a gene Bm-app is responsible for the silkworm minute wing (mw) mutation which exhibits exceedingly small wings during pupal and adult stages. Compared with the wild type silkworm, relative messenger RNA expression of Bm-app is significantly decreased in the u11 mutant strain which shows mw phenotype. A 10 bp insertion in the putative promoter region of the Bm-app gene in mw mutant strain was identified and the dual luciferase assay revealed that this insertion decreased Bm-app promoter activity. Furthermore, clustered regularly interspaced short palindromic repeats/RNA-guided Cas9 nucleases-mediated depletion of the Bm-app induced similar wing defects which appeared in the mw mutant, demonstrating that Bm-app controls wing development in B. mori. Bm-app encodes a palmitoyltransferase and is responsible for the palmitoylation of selected cytoplasmic proteins, indicating that it is required for cell mitosis and growth during wing development. We also discuss the possibility that Bm-app regulates wing development through the Hippo signaling pathway in B. mori.
Keywords: Bombyx mori; CRISPR/Cas9; minute wing; palmitoyltransferase; promoter.
© 2018 Institute of Zoology, Chinese Academy of Sciences.
Figures





Similar articles
-
BmSd gene regulates the silkworm wing size by affecting the Hippo pathway.Insect Sci. 2020 Aug;27(4):655-664. doi: 10.1111/1744-7917.12702. Epub 2019 Jul 21. Insect Sci. 2020. PMID: 31225693
-
BmBR-C Z4 is an upstream regulatory factor of BmPOUM2 controlling the pupal specific expression of BmWCP4 in the silkworm, Bombyx mori.Insect Biochem Mol Biol. 2015 Nov;66:42-50. doi: 10.1016/j.ibmb.2015.09.006. Epub 2015 Sep 10. Insect Biochem Mol Biol. 2015. PMID: 26363295
-
BR-C Z4 and FoxJ interact to regulate expression of a chitin synthase gene CHSA-2b in the pupal wing discs of the silkworm, Bombyx mori.Insect Biochem Mol Biol. 2020 Jan;116:103264. doi: 10.1016/j.ibmb.2019.103264. Epub 2019 Nov 7. Insect Biochem Mol Biol. 2020. PMID: 31707207
-
Transcriptional regulation of cuticular genes during insect metamorphosis.Front Biosci (Landmark Ed). 2020 Jan 1;25(1):106-117. doi: 10.2741/4796. Front Biosci (Landmark Ed). 2020. PMID: 31585879 Review.
-
Recent progress in genome engineering techniques in the silkworm, Bombyx mori.Dev Growth Differ. 2014 Jan;56(1):14-25. doi: 10.1111/dgd.12096. Epub 2013 Nov 1. Dev Growth Differ. 2014. PMID: 24175911 Review.
Cited by
-
Gtsf1 is essential for proper female sex determination and transposon silencing in the silkworm, Bombyx mori.PLoS Genet. 2020 Nov 2;16(11):e1009194. doi: 10.1371/journal.pgen.1009194. eCollection 2020 Nov. PLoS Genet. 2020. PMID: 33137136 Free PMC article.
-
CRISPR/Cas Technology in Insect Insecticide Resistance.Insects. 2025 Mar 26;16(4):345. doi: 10.3390/insects16040345. Insects. 2025. PMID: 40332816 Free PMC article. Review.
-
Bmapaf-1 is Involved in the Response against BmNPV Infection by the Mitochondrial Apoptosis Pathway.Insects. 2020 Sep 22;11(9):647. doi: 10.3390/insects11090647. Insects. 2020. PMID: 32971727 Free PMC article.
-
Advances in Editing Silkworms (Bombyx mori) Genome by Using the CRISPR-Cas System.Insects. 2021 Dec 27;13(1):28. doi: 10.3390/insects13010028. Insects. 2021. PMID: 35055871 Free PMC article. Review.
-
Bombyx mori Vps13d is a key gene affecting silk yield.PLoS One. 2022 Jul 7;17(7):e0270840. doi: 10.1371/journal.pone.0270840. eCollection 2022. PLoS One. 2022. PMID: 35797274 Free PMC article.
References
-
- Bennett, F.C. and Harvey, K.F. (2006) Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Current Biology, 16, 2101–2110. - PubMed
-
- Cao, G.L. , Gong, Y.C. , Hu, X.L. , Zhu, M. , Liang, Z. , Huang, L.X . et al (2018) Identification of tarsal‐less peptides from the silkworm Bombyx mori . Applied Microbiology and Biotechnology, 102, 1809–1822. - PubMed
-
- Cho, E. , Feng, Y. , Rauskolb, C. , Maitra, S. , Fehon, R. and Irvine, K.D. (2006) Delineation of a Fat tumor suppressor pathway. Nature Genetics, 38, 1142–1150. - PubMed
-
- Drisdel, R.C. , Alexander, J.K. , Sayeed, A. and Green, W.N. (2006) Assays of protein palmitoylation. Methods, 40,127–134. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

