Genome analysis of the yeast Diutina catenulata, a member of the Debaryomycetaceae/Metschnikowiaceae (CTG-Ser) clade
- PMID: 29944657
- PMCID: PMC6019693
- DOI: 10.1371/journal.pone.0198957
Genome analysis of the yeast Diutina catenulata, a member of the Debaryomycetaceae/Metschnikowiaceae (CTG-Ser) clade
Abstract
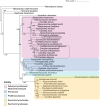
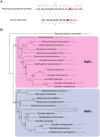
Diutina catenulata (Candida catenulata) is an ascomycetous yeast that has been isolated from humans, animals and environmental sources. The species is a contaminant of dairy products, and has been linked to superficial and invasive infections in both humans and animals. Previous phylogenetic analyses have assigned the species to the Saccharomycetales, but failed to identify its specific clade. Here, we report the genome sequence of an environmental isolate of D. catenulata. Examination of the tRNA repertoire and coding potential of this species shows that it translates the CUG codon as serine and not leucine. In addition, two phylogenetic analyses using 204 ubiquitous gene family alignments and 3,826 single-copy genes both confirm the placement of the species in the Debaryomycetaceae/Metschnikowiaceae, or CTG-Ser clade. The sequenced isolate contains an MTLα idiomorph. However, unlike most MTL loci in related species, poly (A) polymerase (PAP) is not adjacent to MTLα1.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Delavenne E, Mounier J, Asmani K, Jany JL, Barbier G, Le Blay G (2011) Fungal diversity in cow, goat and ewe milk. Int J Food Microbiol 151: 247–251. doi: 10.1016/j.ijfoodmicro.2011.08.029 - DOI - PubMed
-
- Gkatzionis K, Yunita D, Linforth RS, Dickinson M, Dodd CE (2014) Diversity and activities of yeasts from different parts of a Stilton cheese. Int J Food Microbiol 177: 109–116. doi: 10.1016/j.ijfoodmicro.2014.02.016 - DOI - PubMed
-
- Cogan TM, Goerges S, Gelsomino R, Larpin S, Hohenegger M, Bora N, et al. (2014) Biodiversity of the surface microbial consortia from Limburger, Reblochon, Livarot, Tilsit, and Gubbeen cheeses. Microbiol Spectr 2: CM-0010-2012. - PubMed
-
- Santin R, Mattei AS, Waller SB, Madrid IM, Cleff MB, Xavier MO, et al. (2013) Clinical and mycological analysis of dog’s oral cavity. Braz J Microbiol 44: 139–143. doi: 10.1590/S1517-83822013005000018 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

