Transformative Opportunities for Single-Cell Proteomics
- PMID: 29945450
- PMCID: PMC6089608
- DOI: 10.1021/acs.jproteome.8b00257
Transformative Opportunities for Single-Cell Proteomics
Abstract
Many pressing medical challenges, such as diagnosing disease, enhancing directed stem-cell differentiation, and classifying cancers, have long been hindered by limitations in our ability to quantify proteins in single cells. Mass spectrometry (MS) is poised to transcend these limitations by developing powerful methods to routinely quantify thousands of proteins and proteoforms across many thousands of single cells. We outline specific technological developments and ideas that can increase the sensitivity and throughput of single-cell MS by orders of magnitude and usher in this new age. These advances will transform medicine and ultimately contribute to understanding biological systems on an entirely new level.
Keywords: Simpson’s paradox; causal inference; counting noise; disease diagnosis; network inference; sample preparation; single-cell analysis; single-cell mass-spectrometry; systems biology; ultrasensitive proteomics.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
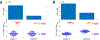

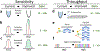
References
-
- Chalfie M; Tu Y; Euskirchen G; Ward WW; Prasher DC Green fluorescent protein as a marker for gene expression. Science 1994, 263, 802–805. - PubMed
-
- Caprioli RM; Farmer TB; Gile J Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal. Chem 1997, 69, 4751–4760. - PubMed
-
- Bandura DR; et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem 2009, 81, 6813–6822. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

