Gene promoter and exon DNA methylation changes in colon cancer development - mRNA expression and tumor mutation alterations
- PMID: 29945573
- PMCID: PMC6020382
- DOI: 10.1186/s12885-018-4609-x
Gene promoter and exon DNA methylation changes in colon cancer development - mRNA expression and tumor mutation alterations
Abstract
Background: DNA mutations occur randomly and sporadically in growth-related genes, mostly on cytosines. Demethylation of cytosines may lead to genetic instability through spontaneous deamination. Aims were whole genome methylation and targeted mutation analysis of colorectal cancer (CRC)-related genes and mRNA expression analysis of TP53 pathway genes.
Methods: Long interspersed nuclear element-1 (LINE-1) BS-PCR followed by pyrosequencing was performed for the estimation of global DNA metlyation levels along the colorectal normal-adenoma-carcinoma sequence. Methyl capture sequencing was done on 6 normal adjacent (NAT), 15 adenomatous (AD) and 9 CRC tissues. Overall quantitative methylation analysis, selection of top hyper/hypomethylated genes, methylation analysis on mutation regions and TP53 pathway gene promoters were performed. Mutations of 12 CRC-related genes (APC, BRAF, CTNNB1, EGFR, FBXW7, KRAS, NRAS, MSH6, PIK3CA, SMAD2, SMAD4, TP53) were evaluated. mRNA expression of TP53 pathway genes was also analyzed.
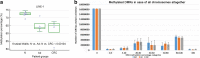
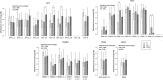
Results: According to the LINE-1 methylation results, overall hypomethylation was observed along the normal-adenoma-carcinoma sequence. Within top50 differential methylated regions (DMRs), in AD-N comparison TP73, NGFR, PDGFRA genes were hypermethylated, FMN1, SLC16A7 genes were hypomethylated. In CRC-N comparison DKK2, SDC2, SOX1 genes showed hypermethylation, while ERBB4, CREB5, CNTN1 genes were hypomethylated. In certain mutation hot spot regions significant DNA methylation alterations were detected. The TP53 gene body was addressed by hypermethylation in adenomas. APC, TP53 and KRAS mutations were found in 30, 15, 21% of adenomas, and in 29, 53, 29% of CRCs, respectively. mRNA expression changes were observed in several TP53 pathway genes showing promoter methylation alterations.
Conclusions: DNA methylation with consecutive phenotypic effect can be observed in a high number of promoter and gene body regions through CRC development.
Keywords: Adenoma; Colorectal cancer; DNA methylation; Methyl capture sequencing; Mutation; TP53 signaling pathway.
Conflict of interest statement
Ethics approval and consent to participate
The study was conducted according to the Helsinki declaration and approved by the local ethics committee and government authorities (Regional and Institutional Committee of Science and Research Ethics (TUKEB) Nr.: 69/2008, 202/2009 and 23,970/2011 Semmelweis University, Budapest, Hungary). All routine colonic biopsy samples from the patients were taken after written informed consent and ethical permission was obtained for participation in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

