Altering cortical input unmasks synaptic phenotypes in the YAC128 cortico-striatal co-culture model of Huntington disease
- PMID: 29945611
- PMCID: PMC6020351
- DOI: 10.1186/s12915-018-0526-3
Altering cortical input unmasks synaptic phenotypes in the YAC128 cortico-striatal co-culture model of Huntington disease
Abstract
Background: Huntington disease (HD) is a fatal neurodegenerative disorder caused by a CAG expansion in the huntingtin (HTT) gene, leading to selective and progressive neuronal death predominantly in the striatum. Mutant HTT expression causes dysfunctional cortico-striatal (CS) transmission, loss of CS synapses, and striatal medium spiny neuron (MSN) dendritic spine instability prior to neuronal death. Co-culturing cortical and striatal neurons in vitro promotes the formation of functional CS synapses and is a widely used approach to elucidate pathogenic mechanisms of HD and to validate potential synapto-protective therapies. A number of relevant in vivo synaptic phenotypes from the YAC128 HD mouse model, which expresses full-length transgenic human mutant HTT, are recapitulated in CS co-culture by 21 days in vitro (DIV). However, striatal spine loss, which occurs in HD patients and in vivo animal models, has been observed in YAC128 CS co-culture in some studies but not in others, leading to difficulties in reproducing and interpreting results. Here, we investigated whether differences in the relative proportion of cortical and striatal neurons alter YAC128 synaptic phenotypes in this model.
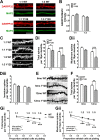
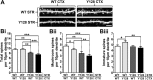
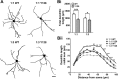
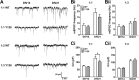
Results: YAC128 MSNs in 1:1 CS co-culture exhibited impaired dendritic length and complexity compared to wild-type, whereas reducing cortical input using a 1:3 CS ratio revealed a dramatic loss of YAC128 MSN dendritic spines. Chimeric experiments determined that this spine instability was primarily cell autonomous, depending largely on mutant HTT expression in striatal neurons. Moreover, we found that spontaneous electrophysiological MSN activity correlated closely with overall dendritic length, with no differences observed between genotypes in 1:3 co-cultures despite significant YAC128 spine loss. Finally, limiting cortical input with a 1:3 CS ratio impaired the basal survival of YAC128 neurons at DIV21, and this was partially selective for dopamine- and cAMP-regulated phosphoprotein 32-positive MSNs.
Conclusions: Our findings reconcile previous discordant reports of spine loss in this model, and improve the utility and reliability of the CS co-culture for the development of novel therapeutic strategies for HD.
Keywords: Huntington disease; YAC128, DARPP32; corticostriatal co-culture; dendrite; huntingtin; spine; synapse.
Conflict of interest statement
Ethics approval and consent to participate
All experiments were performed according to protocols approved by the University of British Columbia Animal Care Committee (Protocol number A16-0130).
Competing interests
MRH was an employee of Teva Pharmaceuticals, Inc. during this study. Teva did not play a role in the design, collection, analysis, or interpretation of data in this study. All other authors declare no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Comment in
-
Synaptopathy, circuitopathy and the computational biology of Huntington's disease.BMC Biol. 2018 Jun 27;16(1):71. doi: 10.1186/s12915-018-0539-y. BMC Biol. 2018. PMID: 29945620 Free PMC article.
Similar articles
-
Enhanced Store-Operated Calcium Entry Leads to Striatal Synaptic Loss in a Huntington's Disease Mouse Model.J Neurosci. 2016 Jan 6;36(1):125-41. doi: 10.1523/JNEUROSCI.1038-15.2016. J Neurosci. 2016. PMID: 26740655 Free PMC article.
-
Mitigation of augmented extrasynaptic NMDAR signaling and apoptosis in cortico-striatal co-cultures from Huntington's disease mice.Neurobiol Dis. 2012 Oct;48(1):40-51. doi: 10.1016/j.nbd.2012.05.013. Epub 2012 Jun 2. Neurobiol Dis. 2012. PMID: 22668780
-
Full length mutant huntingtin is required for altered Ca2+ signaling and apoptosis of striatal neurons in the YAC mouse model of Huntington's disease.Neurobiol Dis. 2008 Jul;31(1):80-8. doi: 10.1016/j.nbd.2008.03.010. Epub 2008 Apr 16. Neurobiol Dis. 2008. PMID: 18502655 Free PMC article.
-
Molecular insights into cortico-striatal miscommunications in Huntington's disease.Curr Opin Neurobiol. 2018 Feb;48:79-89. doi: 10.1016/j.conb.2017.10.019. Epub 2017 Nov 7. Curr Opin Neurobiol. 2018. PMID: 29125980 Free PMC article. Review.
-
Cell-Autonomous and Non-cell-Autonomous Pathogenic Mechanisms in Huntington's Disease: Insights from In Vitro and In Vivo Models.Neurotherapeutics. 2019 Oct;16(4):957-978. doi: 10.1007/s13311-019-00782-9. Neurotherapeutics. 2019. PMID: 31529216 Free PMC article. Review.
Cited by
-
BDNF and TRiC-inspired reagent rescue cortical synaptic deficits in a mouse model of Huntington's disease.Neurobiol Dis. 2024 Jun 1;195:106502. doi: 10.1016/j.nbd.2024.106502. Epub 2024 Apr 10. Neurobiol Dis. 2024. PMID: 38608784 Free PMC article.
-
C57BL/6 Background Attenuates mHTT Toxicity in the Striatum of YAC128 Mice.Int J Mol Sci. 2021 Nov 23;22(23):12664. doi: 10.3390/ijms222312664. Int J Mol Sci. 2021. PMID: 34884469 Free PMC article.
-
Structural Plasticity of the Hippocampus in Neurodegenerative Diseases.Int J Mol Sci. 2022 Mar 20;23(6):3349. doi: 10.3390/ijms23063349. Int J Mol Sci. 2022. PMID: 35328770 Free PMC article. Review.
-
The Interaction of Aging and Cellular Stress Contributes to Pathogenesis in Mouse and Human Huntington Disease Neurons.Front Aging Neurosci. 2020 Sep 18;12:524369. doi: 10.3389/fnagi.2020.524369. eCollection 2020. Front Aging Neurosci. 2020. PMID: 33192449 Free PMC article.
-
Soluble TREM2 ameliorates pathological phenotypes in ischemic stroke models via modulating neuronal and microglial functions.Exp Brain Res. 2025 May 16;243(6):149. doi: 10.1007/s00221-025-07094-9. Exp Brain Res. 2025. PMID: 40379866
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

