Actin polymerization controls cilia-mediated signaling
- PMID: 29945904
- PMCID: PMC6122990
- DOI: 10.1083/jcb.201703196
Actin polymerization controls cilia-mediated signaling
Abstract
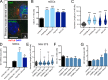
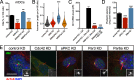
Primary cilia are polarized organelles that allow detection of extracellular signals such as Hedgehog (Hh). How the cytoskeleton supporting the cilium generates and maintains a structure that finely tunes cellular response remains unclear. Here, we find that regulation of actin polymerization controls primary cilia and Hh signaling. Disrupting actin polymerization, or knockdown of N-WASp/Arp3, increases ciliation frequency, axoneme length, and Hh signaling. Cdc42, a potent actin regulator, recruits both atypical protein pinase C iota/lambda (aPKC) and Missing-in-Metastasis (MIM) to the basal body to maintain actin polymerization and restrict axoneme length. Transcriptome analysis implicates the Src pathway as a major aPKC effector. aPKC promotes whereas MIM antagonizes Src activity to maintain proper levels of primary cilia, actin polymerization, and Hh signaling. Hh pathway activation requires Smoothened-, Gli-, and Gli1-specific activation by aPKC. Surprisingly, longer axonemes can amplify Hh signaling, except when aPKC is disrupted, reinforcing the importance of the Cdc42-aPKC-Gli axis in actin-dependent regulation of primary cilia signaling.
© 2018 Drummond et al.
Figures




Similar articles
-
GLI activation by atypical protein kinase C ι/λ regulates the growth of basal cell carcinomas.Nature. 2013 Feb 28;494(7438):484-8. doi: 10.1038/nature11889. Nature. 2013. PMID: 23446420 Free PMC article.
-
MIM and cortactin antagonism regulates ciliogenesis and hedgehog signaling.Dev Cell. 2010 Aug 17;19(2):270-83. doi: 10.1016/j.devcel.2010.07.009. Dev Cell. 2010. PMID: 20708589 Free PMC article.
-
The intrahepatic signalling niche of hedgehog is defined by primary cilia positive cells during chronic liver injury.J Hepatol. 2014 Jan;60(1):143-51. doi: 10.1016/j.jhep.2013.08.012. Epub 2013 Aug 23. J Hepatol. 2014. PMID: 23978713
-
Developmental and regenerative paradigms of cilia regulated hedgehog signaling.Semin Cell Dev Biol. 2021 Feb;110:89-103. doi: 10.1016/j.semcdb.2020.05.029. Epub 2020 Jun 12. Semin Cell Dev Biol. 2021. PMID: 32540122 Free PMC article. Review.
-
Cdc42 in actin dynamics: An ordered pathway governed by complex equilibria and directional effector handover.Small GTPases. 2017 Oct 2;8(4):237-244. doi: 10.1080/21541248.2016.1215657. Epub 2016 Aug 11. Small GTPases. 2017. PMID: 27715449 Free PMC article. Review.
Cited by
-
Sequential events during the quiescence to proliferation transition establish patterns of follicle cell differentiation in the Drosophila ovary.Biol Open. 2023 Jan 1;12(1):bio059625. doi: 10.1242/bio.059625. Epub 2023 Jan 12. Biol Open. 2023. PMID: 36524613 Free PMC article.
-
BAR domain proteins-a linkage between cellular membranes, signaling pathways, and the actin cytoskeleton.Biophys Rev. 2018 Dec;10(6):1587-1604. doi: 10.1007/s12551-018-0467-7. Epub 2018 Nov 19. Biophys Rev. 2018. PMID: 30456600 Free PMC article. Review.
-
BAG3 regulates cilia homeostasis of glioblastoma via its WW domain.Biofactors. 2024 Nov-Dec;50(6):1113-1133. doi: 10.1002/biof.2060. Epub 2024 Apr 24. Biofactors. 2024. PMID: 38655699 Free PMC article.
-
Regulation of the length of neuronal primary cilia and its potential effects on signalling.Trends Cell Biol. 2023 Nov;33(11):979-990. doi: 10.1016/j.tcb.2023.05.005. Epub 2023 Jun 10. Trends Cell Biol. 2023. PMID: 37302961 Free PMC article. Review.
-
The Roles of Primary Cilia in Cardiovascular Diseases.Cells. 2018 Nov 27;7(12):233. doi: 10.3390/cells7120233. Cells. 2018. PMID: 30486394 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

