Genomic Selection in Preliminary Yield Trials in a Winter Wheat Breeding Program
- PMID: 29945967
- PMCID: PMC6071594
- DOI: 10.1534/g3.118.200415
Genomic Selection in Preliminary Yield Trials in a Winter Wheat Breeding Program
Abstract
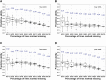
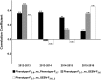
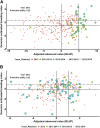
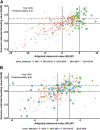
Genomic prediction (GP) is now routinely performed in crop plants to predict unobserved phenotypes. The use of predicted phenotypes to make selections is an active area of research. Here, we evaluate GP for predicting grain yield and compare genomic and phenotypic selection by tracking lines advanced. We examined four independent nurseries of F3:6 and F3:7 lines trialed at 6 to 10 locations each year. Yield was analyzed using mixed models that accounted for experimental design and spatial variations. Genotype-by-sequencing provided nearly 27,000 high-quality SNPs. Average genomic predictive ability, estimated for each year by randomly masking lines as missing in steps of 10% from 10 to 90%, and using the remaining lines from the same year as well as lines from other years in a training set, ranged from 0.23 to 0.55. The predictive ability estimated for a new year using the other years ranged from 0.17 to 0.28. Further, we tracked lines advanced based on phenotype from each of the four F3:6 nurseries. Lines with both above average genomic estimated breeding value (GEBV) and phenotypic value (BLUP) were retained for more years compared to lines with either above average GEBV or BLUP alone. The number of lines selected for advancement was substantially greater when predictions were made with 50% of the lines from the testing year added to the training set. Hence, evaluation of only 50% of the lines yearly seems possible. This study provides insights to assess and integrate genomic selection in breeding programs of autogamous crops.
Keywords: GenPred; Genomic selection; Triticum aestivum; genomic best linear unbiased prediction; genomic prediction; genotyping-by-sequencing; shared data resources; spatial variation.
Copyright © 2018 Belamkar et al.
Figures

References
-
- Arruda M. P., Lipka A. E., Brown P. J., Krill A. M., Thurber C., et al. , 2016. Comparing genomic selection and marker-assisted selection for Fusarium head blight resistance in wheat (Triticum aestivum L.). Mol. Breed. 36: 84.
-
- Asoro F. G., Newell M. A., Beavis W. D., Scott M. P., Jannink J.-L., 2011. Accuracy and training population design for genomic selection on quantitative traits in elite North American oats. Plant Genome 4: 132–144. 10.3835/plantgenome2011.02.0007 - DOI
-
- Asoro F. G., Newell M. A., Beavis W. D., Scott M. P., Tinker N. A., et al. , 2013. Genomic, Marker-Assisted, and Pedigree-BLUP Selection Methods for β-Glucan Concentration in Elite Oat. Crop Sci. 53: 1894–1906. 10.2135/cropsci2012.09.0526 - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
