Evolutionary genomic dynamics of Peruvians before, during, and after the Inca Empire
- PMID: 29946025
- PMCID: PMC6048481
- DOI: 10.1073/pnas.1720798115
Evolutionary genomic dynamics of Peruvians before, during, and after the Inca Empire
Abstract
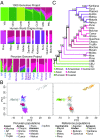
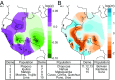
Native Americans from the Amazon, Andes, and coastal geographic regions of South America have a rich cultural heritage but are genetically understudied, therefore leading to gaps in our knowledge of their genomic architecture and demographic history. In this study, we sequence 150 genomes to high coverage combined with an additional 130 genotype array samples from Native American and mestizo populations in Peru. The majority of our samples possess greater than 90% Native American ancestry, which makes this the most extensive Native American sequencing project to date. Demographic modeling reveals that the peopling of Peru began ∼12,000 y ago, consistent with the hypothesis of the rapid peopling of the Americas and Peruvian archeological data. We find that the Native American populations possess distinct ancestral divisions, whereas the mestizo groups were admixtures of multiple Native American communities that occurred before and during the Inca Empire and Spanish rule. In addition, the mestizo communities also show Spanish introgression largely following Peruvian Independence, nearly 300 y after Spain conquered Peru. Further, we estimate migration events between Peruvian populations from all three geographic regions with the majority of between-region migration moving from the high Andes to the low-altitude Amazon and coast. As such, we present a detailed model of the evolutionary dynamics which impacted the genomes of modern-day Peruvians and a Native American ancestry dataset that will serve as a beneficial resource to addressing the underrepresentation of Native American ancestry in sequencing studies.
Keywords: Native American demography; fine-scale structure; gene flow; identity by descent; population history.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

