Activation of the p53-MDM4 regulatory axis defines the anti-tumour response to PRMT5 inhibition through its role in regulating cellular splicing
- PMID: 29946150
- PMCID: PMC6018746
- DOI: 10.1038/s41598-018-28002-y
Activation of the p53-MDM4 regulatory axis defines the anti-tumour response to PRMT5 inhibition through its role in regulating cellular splicing
Abstract
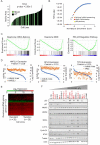
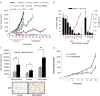
Evasion of the potent tumour suppressor activity of p53 is one of the hurdles that must be overcome for cancer cells to escape normal regulation of cellular proliferation and survival. In addition to frequent loss of function mutations, p53 wild-type activity can also be suppressed post-translationally through several mechanisms, including the activity of PRMT5. Here we describe broad anti-proliferative activity of potent, selective, reversible inhibitors of protein arginine methyltransferase 5 (PRMT5) including GSK3326595 in human cancer cell lines representing both hematologic and solid malignancies. Interestingly, PRMT5 inhibition activates the p53 pathway via the induction of alternative splicing of MDM4. The MDM4 isoform switch and subsequent p53 activation are critical determinants of the response to PRMT5 inhibition suggesting that the integrity of the p53-MDM4 regulatory axis defines a subset of patients that could benefit from treatment with GSK3326595.
Conflict of interest statement
SVG, WAK, CT, MBP, X-PZ, RMdO, BL, MM, CC, NJ, RK, OB are past or present employees and stakeholders of GlaxoSmithKline. EP, KD, AB, and CM are past or present employees and stakeholders of Epizyme.
Figures
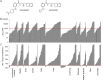
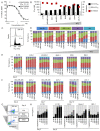
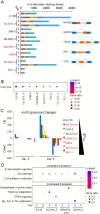

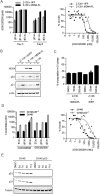
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

