Differentially methylated regions in T cells identify kidney transplant patients at risk for de novo skin cancer
- PMID: 29946375
- PMCID: PMC6006560
- DOI: 10.1186/s13148-018-0519-7
Differentially methylated regions in T cells identify kidney transplant patients at risk for de novo skin cancer
Abstract
Background: Cutaneous squamous cell carcinoma (cSCC) occurs 65-200 times more in immunosuppressed organ transplant patients than in the general population. T cells, which are targeted by the given immunosuppressive drugs, are involved in anti-tumor immune surveillance and are functionally regulated by DNA methylation. Prior to kidney transplantation, we aim to discover differentially methylated regions (DMRs) in T cells involved in de novo post-transplant cSCC development.
Methods: We matched 27 kidney transplant patients with a future de novo cSCC after transplantation to 27 kidney transplant patients without cSCC and studied genome-wide DNA methylation of T cells prior to transplantation. From 11 out of the 27 cSCC patients, the DNA methylation of T cells after transplantation was also examined to assess stability of the observed differences in DNA methylation. Raw methylation values obtained with the 450k array were confirmed with pyrosequencing.
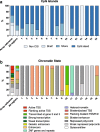
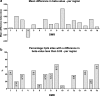
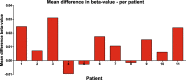
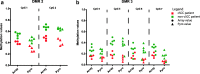
Results: We found 16 DMRs between patients with a future cSCC and those who do not develop this complication after transplantation. The majority of the DMRs were located in regulatory genomic regions such as flanking bivalent transcription start sites and bivalent enhancer regions, and most of the DMRs contained CpG islands. Examples of genes annotated to the DMRs are ZNF577, coding for a zinc-finger protein, and FLOT1, coding for a protein involved in T cell migration. The longitudinal analysis revealed that DNA methylation of 9 DMRs changed significantly after transplantation. DNA methylation of 5 out of 16 DMRs was relatively stable, with a variation in beta-value lower than 0.05 for at least 50% of the CpG sites within that region.
Conclusions: This is the first study demonstrating that DNA methylation of T cells from patients with a future de novo post-transplant cSCC is different from patients without cSCC. These results were obtained before transplantation, a clinically relevant time point for cSCC risk assessment. Several DNA methylation profiles remained relatively stable after transplantation, concluding that these are minimally affected by the transplantation and possibly have a lasting effect on post-transplant cSCC development.
Keywords: Cutaneous squamous cell carcinoma; DNA methylation; Epigenetics; Non-melanoma skin cancer; Solid organ transplantation; T lymphocytes.
Conflict of interest statement
Anonymized biobank samples were used in this study; this approach had been approved by the local ethical committee (MEC-2015-642).Not applicableThe authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Krynitz B, Edgren G, Lindelof B, Baecklund E, Brattstrom C, Wilczek H, Smedby KE. Risk of skin cancer and other malignancies in kidney, liver, heart and lung transplant recipients 1970 to 2008—a Swedish population-based study. Int J Cancer. 2013;132(6):1429–1438. doi: 10.1002/ijc.27765. - DOI - PubMed
-
- Euvrard S, Kanitakis J, Decullier E, Butnaru AC, Lefrançois N, Boissonnat P, Sebbag L, Garnier J-L, Pouteil-Noble C, Cahen R, et al. Subsequent skin cancers in kidney and heart transplant recipients after the first squamous cell carcinoma. Transplantation. 2006;81(8):1093–1100. doi: 10.1097/01.tp.0000209921.60305.d9. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

