Influence of isopropylmalate synthase OsIPMS1 on seed vigour associated with amino acid and energy metabolism in rice
- PMID: 29947463
- PMCID: PMC6335077
- DOI: 10.1111/pbi.12979
Influence of isopropylmalate synthase OsIPMS1 on seed vigour associated with amino acid and energy metabolism in rice
Abstract
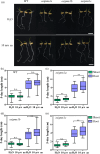
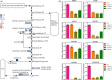
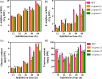
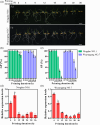
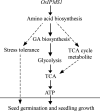
Seed vigour is an imperative trait for the direct seeding of rice. Isopropylmalate synthase (IPMS) catalyses the committed step of leucine (Leu) biosynthesis, but its effect on seed vigour remains unclear. In this study, rice OsIPMS1 and OsIPMS2 was cloned, and the roles of OsIPMS1 in seed vigour were mainly investigated. OsIPMS1 and OsIPMS2 catalyse Leu biosynthesis, and Leu feedback inhibits their IPMS activities. Disruption of OsIPMS1 resulted in low seed vigour under various conditions, which might be tightly associated with the reduction of amino acids in germinating seeds. Eleven amino acids that associated with stress tolerance, GA biosynthesis and tricarboxylic acid (TCA) cycle were significantly reduced in osipms1 mutants compared with those in wide type (WT) during seed germination. Transcriptome analysis indicated that a total of 1209 differentially expressed genes (DEGs) were altered in osipms1a mutant compared with WT at the early germination stage, wherein most of the genes were involved in glycolysis/gluconeogenesis, protein processing, pyruvate, carbon, fructose and mannose metabolism. Further analysis confirmed that the regulation of OsIPMS1 in seed vigour involved in starch hydrolysis, glycolytic activity and energy levels in germinating seeds. The effects of seed priming were tightly associated with the mRNA levels of OsIPMS1 in priming seeds. The OsIPMS1 might be used as a biomarker to determine the best stop time-point of seed priming in rice. This study provides novel insights into the function of OsIPMS1 on seed vigour and should have practical applications in seed priming of rice.
Keywords: Oryza sativa; isopropylmalate synthase; seed priming; seed vigour.
© 2018 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Abe, A. , Takagi, H. , Fujibe, T. , Aya, K. , Kojima, M. , Sakakibara, H. , Uemura, A. et al. (2012) OsGA20ox1, a candidate gene for a major QTL controlling seedling vigor in rice. Theor. Appl. Genet. 125, 647–657. - PubMed
-
- Angelovici, R. , Fait, A. , Fernie, A.R. and Galili, G. (2011) A seed high‐lysine trait is negatively associated with the TCA cycle and slows down Arabidopsis seed germination. New Phytol. 189, 148–159. - PubMed
-
- Araújo, W.L. , Ishizaki, K. , Nunes‐Nesi, A. , Larson, T.R. , Tohge, T. , Krahnert, I. , Witt, S. et al. (2010) Identification of the 2‐hydroxyglutarate and isovaleryl‐CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell, 22, 1549–1563. - PMC - PubMed
-
- Araújo, W.L. , Tohge, T. , Ishizaki, K. , Leaver, C.J. and Fernie, A.R. (2011) Protein degradation ‐ an alternative respiratory substrate for stressed plants. Trends Plant Sci. 16, 489–498. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 31771889/National Natural Science Foundation of China/International
- 2018YFD0100901/National Key Research and Development Plan/International
- 2017KTSCX024/Major Scientific Research Projects of General Colleges and Universities of Guangdong Province/International
- BK20161451/Natural Science Foundation of Jiangsu Province/International
- BK20150675/Natural Science Foundation of Jiangsu Province/International
LinkOut - more resources
Full Text Sources
Other Literature Sources

