Efficient CRISPR/Cas9-mediated editing of trinucleotide repeat expansion in myotonic dystrophy patient-derived iPS and myogenic cells
- PMID: 29947794
- PMCID: PMC6144820
- DOI: 10.1093/nar/gky548
Efficient CRISPR/Cas9-mediated editing of trinucleotide repeat expansion in myotonic dystrophy patient-derived iPS and myogenic cells
Abstract
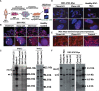
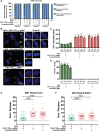
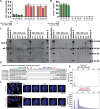
CRISPR/Cas9 is an attractive platform to potentially correct dominant genetic diseases by gene editing with unprecedented precision. In the current proof-of-principle study, we explored the use of CRISPR/Cas9 for gene-editing in myotonic dystrophy type-1 (DM1), an autosomal-dominant muscle disorder, by excising the CTG-repeat expansion in the 3'-untranslated-region (UTR) of the human myotonic dystrophy protein kinase (DMPK) gene in DM1 patient-specific induced pluripotent stem cells (DM1-iPSC), DM1-iPSC-derived myogenic cells and DM1 patient-specific myoblasts. To eliminate the pathogenic gain-of-function mutant DMPK transcript, we designed a dual guide RNA based strategy that excises the CTG-repeat expansion with high efficiency, as confirmed by Southern blot and single molecule real-time (SMRT) sequencing. Correction efficiencies up to 90% could be attained in DM1-iPSC as confirmed at the clonal level, following ribonucleoprotein (RNP) transfection of CRISPR/Cas9 components without the need for selective enrichment. Expanded CTG repeat excision resulted in the disappearance of ribonuclear foci, a quintessential cellular phenotype of DM1, in the corrected DM1-iPSC, DM1-iPSC-derived myogenic cells and DM1 myoblasts. Consequently, the normal intracellular localization of the muscleblind-like splicing regulator 1 (MBNL1) was restored, resulting in the normalization of splicing pattern of SERCA1. This study validates the use of CRISPR/Cas9 for gene editing of repeat expansions.
Figures


Similar articles
-
Comprehensive transcriptome-wide analysis of spliceopathy correction of myotonic dystrophy using CRISPR-Cas9 in iPSCs-derived cardiomyocytes.Mol Ther. 2022 Jan 5;30(1):75-91. doi: 10.1016/j.ymthe.2021.08.004. Epub 2021 Aug 8. Mol Ther. 2022. PMID: 34371182 Free PMC article.
-
CRISPR/Cas9-Induced (CTG⋅CAG)n Repeat Instability in the Myotonic Dystrophy Type 1 Locus: Implications for Therapeutic Genome Editing.Mol Ther. 2017 Jan 4;25(1):24-43. doi: 10.1016/j.ymthe.2016.10.014. Epub 2017 Jan 4. Mol Ther. 2017. PMID: 28129118 Free PMC article.
-
Genome Editing of Expanded CTG Repeats within the Human DMPK Gene Reduces Nuclear RNA Foci in the Muscle of DM1 Mice.Mol Ther. 2019 Aug 7;27(8):1372-1388. doi: 10.1016/j.ymthe.2019.05.021. Epub 2019 Jun 5. Mol Ther. 2019. PMID: 31253581 Free PMC article.
-
Application of CRISPR-Cas9-Mediated Genome Editing for the Treatment of Myotonic Dystrophy Type 1.Mol Ther. 2020 Dec 2;28(12):2527-2539. doi: 10.1016/j.ymthe.2020.10.005. Epub 2020 Oct 14. Mol Ther. 2020. PMID: 33171139 Free PMC article. Review.
-
CRISPR/Cas Applications in Myotonic Dystrophy: Expanding Opportunities.Int J Mol Sci. 2019 Jul 27;20(15):3689. doi: 10.3390/ijms20153689. Int J Mol Sci. 2019. PMID: 31357652 Free PMC article. Review.
Cited by
-
The third generation sequencing: the advanced approach to genetic diseases.Transl Pediatr. 2020 Apr;9(2):163-173. doi: 10.21037/tp.2020.03.06. Transl Pediatr. 2020. PMID: 32477917 Free PMC article. Review.
-
Alternative splicing in neurodegenerative disease and the promise of RNA therapies.Nat Rev Neurosci. 2023 Aug;24(8):457-473. doi: 10.1038/s41583-023-00717-6. Epub 2023 Jun 19. Nat Rev Neurosci. 2023. PMID: 37336982 Review.
-
Improved CRISPR/Cas9 gene editing in primary human myoblasts using low confluency cultures on Matrigel.Skelet Muscle. 2021 Sep 22;11(1):23. doi: 10.1186/s13395-021-00278-1. Skelet Muscle. 2021. PMID: 34551826 Free PMC article.
-
Cas9 editing of ATXN1 in a spinocerebellar ataxia type 1 mice and human iPSC-derived neurons.Mol Ther Nucleic Acids. 2024 Aug 31;35(4):102317. doi: 10.1016/j.omtn.2024.102317. eCollection 2024 Dec 10. Mol Ther Nucleic Acids. 2024. PMID: 39314800 Free PMC article.
-
A CTG repeat-selective chemical screen identifies microtubule inhibitors as selective modulators of toxic CUG RNA levels.Proc Natl Acad Sci U S A. 2019 Oct 15;116(42):20991-21000. doi: 10.1073/pnas.1901893116. Epub 2019 Sep 30. Proc Natl Acad Sci U S A. 2019. PMID: 31570586 Free PMC article.
References
-
- Udd B., Krahe R.. The myotonic dystrophies: molecular, clinical, and therapeutic challenges. Lancet Neurol. 2012; 11:891–905. - PubMed
-
- Theadom A., Rodrigues M., Roxburgh R., Balalla S., Higgins C., Bhattacharjee R., Jones K., Krishnamurthi R., Feigin V.. Prevalence of muscular dystrophies: a systematic literature review. Neuroepidemiology. 2014; 43:259–268. - PubMed
-
- Fu Y.H., Pizzuti A., Fenwick R.G. Jr., King J., Rajnarayan S., Dunne P.W., Dubel J., Nasser G.A., Ashizawa T., de Jong P. et al. . An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 1992; 255:1256–1258. - PubMed
-
- Mahadevan M., Tsilfidis C., Sabourin L., Shutler G., Amemiya C., Jansen G., Neville C., Narang M., Barcelo J., O’Hoy K. et al. . Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science. 1992; 255:1253–1255. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

