The auxin receptor TIR1 homolog (PagFBL 1) regulates adventitious rooting through interactions with Aux/IAA28 in Populus
- PMID: 29949229
- PMCID: PMC6335065
- DOI: 10.1111/pbi.12980
The auxin receptor TIR1 homolog (PagFBL 1) regulates adventitious rooting through interactions with Aux/IAA28 in Populus
Abstract
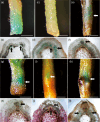
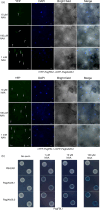
Adventitious roots occur naturally in many species and can also be induced from explants of some tree species including Populus, providing an important means of clonal propagation. Auxin has been identified as playing a crucial role in adventitious root formation, but the associated molecular regulatory mechanisms need to be elucidated. In this study, we examined the role of PagFBL1, the hybrid poplar (Populus alba × P. glandulosa clone 84K) homolog of Arabidopsis auxin receptor TIR1, in adventitious root formation in poplar. Similar to the distribution pattern of auxin during initiation of adventitious roots, PagFBL1 expression was concentrated in the cambium and secondary phloem in stems during adventitious root induction and initiation phases, but decreased in emerging adventitious root primordia. Overexpressing PagFBL1 stimulated adventitious root formation and increased root biomass, while knock-down of PagFBL1 transcript levels delayed adventitious root formation and decreased root biomass. Transcriptome analyses of PagFBL1 overexpressing lines indicated that an extensive remodelling of gene expression was stimulated by auxin signalling pathway during early adventitious root formation. In addition, PagIAA28 was identified as downstream targets of PagFBL1. We propose that the PagFBL1-PagIAA28 module promotes adventitious rooting and could be targeted to improve Populus propagation by cuttings.
Keywords: PagFBL1; PagIAA28; adventitious root development; auxin signalling.
© 2018 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Figures



Similar articles
-
Gibberellins inhibit adventitious rooting in hybrid aspen and Arabidopsis by affecting auxin transport.Plant J. 2014 May;78(3):372-84. doi: 10.1111/tpj.12478. Epub 2014 Apr 2. Plant J. 2014. PMID: 24547703
-
A Molecular Framework for the Control of Adventitious Rooting by TIR1/AFB2-Aux/IAA-Dependent Auxin Signaling in Arabidopsis.Mol Plant. 2019 Nov 4;12(11):1499-1514. doi: 10.1016/j.molp.2019.09.001. Epub 2019 Sep 11. Mol Plant. 2019. PMID: 31520787
-
The cytokinin type-B response regulator PtRR13 is a negative regulator of adventitious root development in Populus.Plant Physiol. 2009 Jun;150(2):759-71. doi: 10.1104/pp.109.137505. Epub 2009 Apr 24. Plant Physiol. 2009. PMID: 19395410 Free PMC article.
-
Unresolved roles of Aux/IAA proteins in auxin responses.Physiol Plant. 2025 Mar-Apr;177(2):e70221. doi: 10.1111/ppl.70221. Physiol Plant. 2025. PMID: 40265222 Free PMC article. Review.
-
Auxin receptors and plant development: a new signaling paradigm.Annu Rev Cell Dev Biol. 2008;24:55-80. doi: 10.1146/annurev.cellbio.23.090506.123214. Annu Rev Cell Dev Biol. 2008. PMID: 18631113 Review.
Cited by
-
Transcriptomic profiles of poplar (Populus simonii × P. nigra) cuttings during adventitious root formation.Front Genet. 2022 Sep 8;13:968544. doi: 10.3389/fgene.2022.968544. eCollection 2022. Front Genet. 2022. PMID: 36160010 Free PMC article.
-
The BTB protein MdBT2 recruits auxin signaling components to regulate adventitious root formation in apple.Plant Physiol. 2022 Jun 1;189(2):1005-1020. doi: 10.1093/plphys/kiac084. Plant Physiol. 2022. PMID: 35218363 Free PMC article.
-
Wound-induced signals regulate root organogenesis in Arabidopsis explants.BMC Plant Biol. 2022 Mar 22;22(1):133. doi: 10.1186/s12870-022-03524-w. BMC Plant Biol. 2022. PMID: 35317749 Free PMC article.
-
Comparative transcriptomic and hormonal analyses reveal potential regulation networks of adventitious root formation in Metasequoia glyptostroboides Hu et Cheng.BMC Genomics. 2024 Nov 18;25(1):1098. doi: 10.1186/s12864-024-10989-6. BMC Genomics. 2024. PMID: 39558286 Free PMC article.
-
Roles of Auxin in the Growth, Development, and Stress Tolerance of Horticultural Plants.Cells. 2022 Sep 5;11(17):2761. doi: 10.3390/cells11172761. Cells. 2022. PMID: 36078168 Free PMC article. Review.
References
-
- Abarca, D. , Pizarro, A. , Hernández, I. , Sánchez, C. , Solana, S.P. , Amo, A.D. , Carneros, E. et al. (2014) The GRAS gene family in pine: transcript expression patterns associated with the maturation‐related decline of competence to form adventitious roots. BMC Plant Biol. 14, 1–19. - PMC - PubMed
-
- Ahkami, A.H. , Melzer, M. , Ghaffari, M.R. , Pollmann, S. , Javid, M.G. , Shahinnia, F. , Hajirezaei, M.R. et al. (2013) Distribution of indole‐3‐acetic acid in petunia hybrida shoot tip cuttings and relationship between auxin transport, carbohydrate metabolism and adventitious root formation. Planta, 238, 499–517. - PMC - PubMed
-
- Ahkami, A. , Scholz, U. , Steuernagel, B. , Strickert, M. , Haensch, K.T. , Druege, U. , Reinhardt, D. et al. (2014) Comprehensive transcriptome analysis unravels the existence of crucial genes regulating primary metabolism during adventitious root formation in petunia hybrida. PLoS ONE, 9, e100997. - PMC - PubMed
-
- Bellamine, J. , Penel, C. , Greppin, H. and Gaspar, T. (1998) Confirmation of the role of auxin and calcium in the late phases of adventitious root formation. Plant Growth Regul. 26, 191–194.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

