Whole-Genome Sequencing in Severe Chronic Obstructive Pulmonary Disease
- PMID: 29949718
- PMCID: PMC6236690
- DOI: 10.1165/rcmb.2018-0088OC
Whole-Genome Sequencing in Severe Chronic Obstructive Pulmonary Disease
Abstract
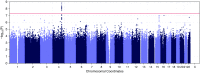
Genome-wide association studies have identified common variants associated with chronic obstructive pulmonary disease (COPD). Whole-genome sequencing (WGS) offers comprehensive coverage of the entire genome, as compared with genotyping arrays or exome sequencing. We hypothesized that WGS in subjects with severe COPD and smoking control subjects with normal pulmonary function would allow us to identify novel genetic determinants of COPD. We sequenced 821 patients with severe COPD and 973 control subjects from the COPDGene and Boston Early-Onset COPD studies, including both non-Hispanic white and African American individuals. We performed single-variant and grouped-variant analyses, and in addition, we assessed the overlap of variants between sequencing- and array-based imputation. Our most significantly associated variant was in a known region near HHIP (combined P = 1.6 × 10-9); additional variants approaching genome-wide significance included previously described regions in CHRNA5, TNS1, and SERPINA6/SERPINA1 (the latter in African American individuals). None of our associations were clearly driven by rare variants, and we found minimal evidence of replication of genes identified by previously reported smaller sequencing studies. With WGS, we identified more than 20 million new variants, not seen with imputation, including more than 10,000 of potential importance in previously identified COPD genome-wide association study regions. WGS in severe COPD identifies a large number of potentially important functional variants, with the strongest associations being in known COPD risk loci, including HHIP and SERPINA1. Larger sample sizes will be needed to identify associated variants in novel regions of the genome.
Trial registration: ClinicalTrials.gov NCT00608764.
Keywords: association studies; chronic obstructive pulmonary disease; whole-genome sequencing.
Figures

Similar articles
-
Genome-Wide Association Analysis of Single-Breath DlCO.Am J Respir Cell Mol Biol. 2019 May;60(5):523-531. doi: 10.1165/rcmb.2018-0384OC. Am J Respir Cell Mol Biol. 2019. PMID: 30694715 Free PMC article. Clinical Trial.
-
Common and Rare Variants Genetic Association Analysis of Cigarettes per Day Among Ever-Smokers in Chronic Obstructive Pulmonary Disease Cases and Controls.Nicotine Tob Res. 2019 May 21;21(6):714-722. doi: 10.1093/ntr/nty095. Nicotine Tob Res. 2019. PMID: 29767774 Free PMC article.
-
Common genetic variants associated with resting oxygenation in chronic obstructive pulmonary disease.Am J Respir Cell Mol Biol. 2014 Nov;51(5):678-87. doi: 10.1165/rcmb.2014-0135OC. Am J Respir Cell Mol Biol. 2014. PMID: 24825563 Free PMC article. Clinical Trial.
-
Applying Functional Genomics to Chronic Obstructive Pulmonary Disease.Ann Am Thorac Soc. 2018 Dec;15(Suppl 4):S239-S242. doi: 10.1513/AnnalsATS.201808-530MG. Ann Am Thorac Soc. 2018. PMID: 30759001 Free PMC article. Review.
-
Hedgehog pathway and its inhibitors in chronic obstructive pulmonary disease (COPD).Pharmacol Ther. 2022 Dec;240:108295. doi: 10.1016/j.pharmthera.2022.108295. Epub 2022 Oct 1. Pharmacol Ther. 2022. PMID: 36191777 Review.
Cited by
-
Update on the Etiology, Assessment, and Management of COPD Cachexia: Considerations for the Clinician.Int J Chron Obstruct Pulmon Dis. 2022 Nov 18;17:2957-2976. doi: 10.2147/COPD.S334228. eCollection 2022. Int J Chron Obstruct Pulmon Dis. 2022. PMID: 36425061 Free PMC article. Review.
-
A flexible and nearly optimal sequential testing approach to randomized testing: QUICK-STOP.Genet Epidemiol. 2020 Mar;44(2):139-147. doi: 10.1002/gepi.22268. Epub 2019 Nov 11. Genet Epidemiol. 2020. PMID: 31713269 Free PMC article.
-
Whole-genome sequencing of African Americans implicates differential genetic architecture in inflammatory bowel disease.Am J Hum Genet. 2021 Mar 4;108(3):431-445. doi: 10.1016/j.ajhg.2021.02.001. Epub 2021 Feb 17. Am J Hum Genet. 2021. PMID: 33600772 Free PMC article.
-
C FTR variants are associated with chronic bronchitis in smokers.Eur Respir J. 2022 Aug 10;60(2):2101994. doi: 10.1183/13993003.01994-2021. Print 2022 Aug. Eur Respir J. 2022. PMID: 34996830 Free PMC article.
-
Network Medicine in the Age of Biomedical Big Data.Front Genet. 2019 Apr 11;10:294. doi: 10.3389/fgene.2019.00294. eCollection 2019. Front Genet. 2019. PMID: 31031797 Free PMC article. Review.
References
-
- Vestbo J, Hurd SS, Agustí AG, Jones PW, Vogelmeier C, Anzueto A, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2013;187:347–365. - PubMed
-
- Ingebrigtsen T, Thomsen SF, Vestbo J, van der Sluis S, Kyvik KO, Silverman EK, et al. Genetic influences on chronic obstructive pulmonary disease – a twin study. Respir Med. 2010;104:1890–1895. - PubMed
-
- Silverman EK, Chapman HA, Drazen JM, Weiss ST, Rosner B, Campbell EJ, et al. Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease: risk to relatives for airflow obstruction and chronic bronchitis. Am J Respir Crit Care Med. 1998;157:1770–1778. - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
- N01 HC095161/HL/NHLBI NIH HHS/United States
- R01 HL077612/HL/NHLBI NIH HHS/United States
- R01 HL071250/HL/NHLBI NIH HHS/United States
- N01 HC095167/HL/NHLBI NIH HHS/United States
- R01 HL089856/HL/NHLBI NIH HHS/United States
- N01 HC095159/HL/NHLBI NIH HHS/United States
- R01 HL135142/HL/NHLBI NIH HHS/United States
- N01 HC095163/HL/NHLBI NIH HHS/United States
- R01 HL071205/HL/NHLBI NIH HHS/United States
- UL1 TR000040/TR/NCATS NIH HHS/United States
- N01 HC095166/HL/NHLBI NIH HHS/United States
- R01 HL113264/HL/NHLBI NIH HHS/United States
- UL1 RR033176/RR/NCRR NIH HHS/United States
- R01 HL089897/HL/NHLBI NIH HHS/United States
- HHSN268201500003C/HL/NHLBI NIH HHS/United States
- N01 HC095168/HL/NHLBI NIH HHS/United States
- R01 HL071251/HL/NHLBI NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- R00 HL121087/HL/NHLBI NIH HHS/United States
- R01 HL071259/HL/NHLBI NIH HHS/United States
- UL1 TR001079/TR/NCATS NIH HHS/United States
- N02 HL064278/HL/NHLBI NIH HHS/United States
- N01 HC095169/HL/NHLBI NIH HHS/United States
- R01 HL093081/HL/NHLBI NIH HHS/United States
- U01 HL089856/HL/NHLBI NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- R01 HL071051/HL/NHLBI NIH HHS/United States
- UL1 TR001420/TR/NCATS NIH HHS/United States
- R01 HL137927/HL/NHLBI NIH HHS/United States
- K01 HL129039/HL/NHLBI NIH HHS/United States
- HHSN268201500003I/HL/NHLBI NIH HHS/United States
- R01 HL117626/HL/NHLBI NIH HHS/United States
- N01 HC095162/HL/NHLBI NIH HHS/United States
- UL1 TR001881/TR/NCATS NIH HHS/United States
- R01 HL131565/HL/NHLBI NIH HHS/United States
- N01 HC095165/HL/NHLBI NIH HHS/United States
- N01 HC095164/HL/NHLBI NIH HHS/United States
- P01 HL132825/HL/NHLBI NIH HHS/United States
- R01 HL071258/HL/NHLBI NIH HHS/United States
- T32 HL007427/HL/NHLBI NIH HHS/United States
- N01 HC095160/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

