Investigating the Promoter of FAT10 Gene in HCC Patients
- PMID: 29949944
- PMCID: PMC6070910
- DOI: 10.3390/genes9070319
Investigating the Promoter of FAT10 Gene in HCC Patients
Abstract
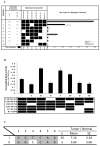
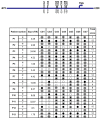
FAT10, which is also known as diubiquitin, has been implicated to play important roles in immune regulation and tumorigenesis. Its expression is up-regulated in the tumors of Hepatocellular Carcinoma (HCC) and other cancer patients. High levels of FAT10 in cells have been shown to result in increased mitotic non-disjunction and chromosome instability, leading to tumorigenesis. To evaluate whether the aberrant up-regulation of the FAT10 gene in the tumors of HCC patients is due to mutations or the aberrant methylation of CG dinucleotides at the FAT10 promoter, sequencing and methylation-specific sequencing of the promoter of FAT10 was performed. No mutations were found that could explain the differential expression of FAT10 between the tumor and non-tumorous tissues of HCC patients. However, six single nucleotide polymorphisms (SNPs), including one that has not been previously reported, were identified at the promoter of the FAT10 gene. Different haplotypes of these SNPs were found to significantly mediate different FAT10 promoter activities. Consistent with the experimental observation, differential FAT10 expression in the tumors of HCC patients carrying haplotype 1 was generally higher than those carrying haplotype II. Notably, the methylation status of this promoter was found to correlate with FAT10 expression levels. Hence, the aberrant overexpression of the FAT10 gene in the tumors of HCC patients is likely due to aberrant methylation, rather than mutations at the FAT10 promoter.
Keywords: FAT10; SNPs; expression; methylation; promoter.
Conflict of interest statement
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources

