Novo&Stitch: accurate reconciliation of genome assemblies via optical maps
- PMID: 29949964
- PMCID: PMC6022655
- DOI: 10.1093/bioinformatics/bty255
Novo&Stitch: accurate reconciliation of genome assemblies via optical maps
Abstract
Motivation: De novo genome assembly is a challenging computational problem due to the high repetitive content of eukaryotic genomes and the imperfections of sequencing technologies (i.e. sequencing errors, uneven sequencing coverage and chimeric reads). Several assembly tools are currently available, each of which has strengths and weaknesses in dealing with the trade-off between maximizing contiguity and minimizing assembly errors (e.g. mis-joins). To obtain the best possible assembly, it is common practice to generate multiple assemblies from several assemblers and/or parameter settings and try to identify the highest quality assembly. Unfortunately, often there is no assembly that both maximizes contiguity and minimizes assembly errors, so one has to compromise one for the other.
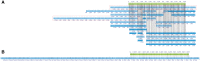
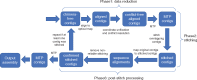
Results: The concept of assembly reconciliation has been proposed as a way to obtain a higher quality assembly by merging or reconciling all the available assemblies. While several reconciliation methods have been introduced in the literature, we have shown in one of our recent papers that none of them can consistently produce assemblies that are better than the assemblies provided in input. Here we introduce Novo&Stitch, a novel method that takes advantage of optical maps to accurately carry out assembly reconciliation (assuming that the assembled contigs are sufficiently long to be reliably aligned to the optical maps, e.g. 50 Kbp or longer). Experimental results demonstrate that Novo&Stitch can double the contiguity (N50) of the input assemblies without introducing mis-joins or reducing genome completeness.
Availability and implementation: Novo&Stitch can be obtained from https://github.com/ucrbioinfo/Novo_Stitch.
Figures
References
-
- Berlin K. et al. (2015) Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol., 33, 623–630. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

