A spectral clustering-based method for identifying clones from high-throughput B cell repertoire sequencing data
- PMID: 29949968
- PMCID: PMC6022594
- DOI: 10.1093/bioinformatics/bty235
A spectral clustering-based method for identifying clones from high-throughput B cell repertoire sequencing data
Abstract
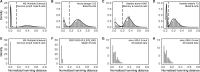
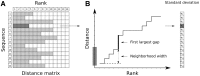
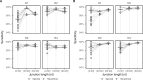
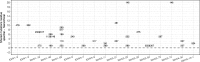
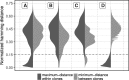
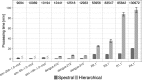
Motivation: B cells derive their antigen-specificity through the expression of Immunoglobulin (Ig) receptors on their surface. These receptors are initially generated stochastically by somatic re-arrangement of the DNA and further diversified following antigen-activation by a process of somatic hypermutation, which introduces mainly point substitutions into the receptor DNA at a high rate. Recent advances in next-generation sequencing have enabled large-scale profiling of the B cell Ig repertoire from blood and tissue samples. A key computational challenge in the analysis of these data is partitioning the sequences to identify descendants of a common B cell (i.e. a clone). Current methods group sequences using a fixed distance threshold, or a likelihood calculation that is computationally-intensive. Here, we propose a new method based on spectral clustering with an adaptive threshold to determine the local sequence neighborhood. Validation using simulated and experimental datasets demonstrates that this method has high sensitivity and specificity compared to a fixed threshold that is optimized for these measures. In addition, this method works on datasets where choosing an optimal fixed threshold is difficult and is more computationally efficient in all cases. The ability to quickly and accurately identify members of a clone from repertoire sequencing data will greatly improve downstream analyses. Clonally-related sequences cannot be treated independently in statistical models, and clonal partitions are used as the basis for the calculation of diversity metrics, lineage reconstruction and selection analysis. Thus, the spectral clustering-based method here represents an important contribution to repertoire analysis.
Availability and implementation: Source code for this method is freely available in the SCOPe (Spectral Clustering for clOne Partitioning) R package in the Immcantation framework: www.immcantation.org under the CC BY-SA 4.0 license.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures




Similar articles
-
Hierarchical Clustering Can Identify B Cell Clones with High Confidence in Ig Repertoire Sequencing Data.J Immunol. 2017 Mar 15;198(6):2489-2499. doi: 10.4049/jimmunol.1601850. Epub 2017 Feb 8. J Immunol. 2017. PMID: 28179494 Free PMC article.
-
Optimized Threshold Inference for Partitioning of Clones From High-Throughput B Cell Repertoire Sequencing Data.Front Immunol. 2018 Jul 26;9:1687. doi: 10.3389/fimmu.2018.01687. eCollection 2018. Front Immunol. 2018. PMID: 30093903 Free PMC article.
-
Somatic hypermutation analysis for improved identification of B cell clonal families from next-generation sequencing data.PLoS Comput Biol. 2020 Jun 23;16(6):e1007977. doi: 10.1371/journal.pcbi.1007977. eCollection 2020 Jun. PLoS Comput Biol. 2020. PMID: 32574157 Free PMC article.
-
The analysis of clonal expansions in normal and autoimmune B cell repertoires.Philos Trans R Soc Lond B Biol Sci. 2015 Sep 5;370(1676):20140239. doi: 10.1098/rstb.2014.0239. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26194753 Free PMC article. Review.
-
Analyzing Immunoglobulin Repertoires.Front Immunol. 2018 Mar 14;9:462. doi: 10.3389/fimmu.2018.00462. eCollection 2018. Front Immunol. 2018. PMID: 29593723 Free PMC article. Review.
Cited by
-
Robust, persistent adaptive immune responses to SARS-CoV-2 in the oropharyngeal lymphoid tissue of children.Res Sq [Preprint]. 2022 Mar 23:rs.3.rs-1276578. doi: 10.21203/rs.3.rs-1276578/v1. Res Sq. 2022. Update in: Nat Immunol. 2023 Jan;24(1):186-199. doi: 10.1038/s41590-022-01367-z. PMID: 35350206 Free PMC article. Updated. Preprint.
-
Single-cell immune repertoire analysis.Nat Methods. 2024 May;21(5):777-792. doi: 10.1038/s41592-024-02243-4. Epub 2024 Apr 18. Nat Methods. 2024. PMID: 38637691 Review.
-
Systemic 4-1BB stimulation augments extrafollicular memory B cell formation and recall responses during Plasmodium infection.Cell Rep. 2025 Apr 22;44(4):115528. doi: 10.1016/j.celrep.2025.115528. Epub 2025 Apr 11. Cell Rep. 2025. PMID: 40215168 Free PMC article.
-
Age-associated B cells are heterogeneous and dynamic drivers of autoimmunity in mice.J Exp Med. 2023 May 1;220(5):e20221346. doi: 10.1084/jem.20221346. Epub 2023 Feb 24. J Exp Med. 2023. PMID: 36828389 Free PMC article.
-
VDJbase: an adaptive immune receptor genotype and haplotype database.Nucleic Acids Res. 2020 Jan 8;48(D1):D1051-D1056. doi: 10.1093/nar/gkz872. Nucleic Acids Res. 2020. PMID: 31602484 Free PMC article.
References
-
- Bannard O., Cyster J.G. (2017) Germinal centers: programmed for affinity maturation and antibody diversification. Curr. Opin. Immunol., 45, 21–30. - PubMed
-
- Boyd S.D., Joshi S.A. (2015) High-throughput DNA sequencing analysis of antibody repertoires In: Crowe,J. (eds) Antibodies for Infectious Diseases. American Society of Microbiology, Washington, DC, pp. 345–362.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

