A space and time-efficient index for the compacted colored de Bruijn graph
- PMID: 29949982
- PMCID: PMC6022659
- DOI: 10.1093/bioinformatics/bty292
A space and time-efficient index for the compacted colored de Bruijn graph
Abstract
Motivation: Indexing reference sequences for search-both individual genomes and collections of genomes-is an important building block for many sequence analysis tasks. Much work has been dedicated to developing full-text indices for genomic sequences, based on data structures such as the suffix array, the BWT and the FM-index. However, the de Bruijn graph, commonly used for sequence assembly, has recently been gaining attention as an indexing data structure, due to its natural ability to represent multiple references using a graphical structure, and to collapse highly-repetitive sequence regions. Yet, much less attention has been given as to how to best index such a structure, such that queries can be performed efficiently and memory usage remains practical as the size and number of reference sequences being indexed grows large.
Results: We present a novel data structure for representing and indexing the compacted colored de Bruijn graph, which allows for efficient pattern matching and retrieval of the reference information associated with each k-mer. As the popularity of the de Bruijn graph as an index has increased over the past few years, so have the number of proposed representations of this structure. Existing structures typically fall into two categories; those that are hashing-based and provide very fast access to the underlying k-mer information, and those that are space-frugal and provide asymptotically efficient but practically slower pattern search. Our representation achieves a compromise between these two extremes. By building upon minimum perfect hashing and making use of succinct representations where applicable, our data structure provides practically fast lookup while greatly reducing the space compared to traditional hashing-based implementations. Further, we describe a sampling scheme for this index, which provides the ability to trade off query speed for a reduction in the index size. We believe this representation strikes a desirable balance between speed and space usage, and allows for fast search on large reference sequences. Finally, we describe an application of this index to the taxonomic read assignment problem. We show that by adopting, essentially, the approach of Kraken, but replacing k-mer presence with coverage by chains of consistent unique maximal matches, we can improve the space, speed and accuracy of taxonomic read assignment.
Availability and implementation: pufferfish is written in C++11, is open source, and is available at https://github.com/COMBINE-lab/pufferfish.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
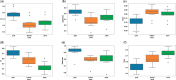
References
-
- Almodaresi F. et al. (2017) Rainbowfish: a succinct colored de Bruijn graph representation. In: Schwartz, R. and Reinert, K. (eds) 17th International Workshop on Algorithms in Bioinformatics (WABI 2017), volume 88 of Leibniz International Proceedings in Informatics (LIPIcs), pp. 18: 1–18: 15, Dagstuhl, Germany. Schloss Dagstuhl–Leibniz-Zentrum fuer Informatik.
-
- Belazzougui D. et al. (2016) Fully dynamic de Bruijn graphs In International Symposium on String Processing and Information Retrieval, Springer, pp. 145–152.
-
- Bowe A. et al. (2012) Succinct de Bruijn graphs. In Proceedings of the International Workshop on Algorithms in Bioinformatics, Springer, pp. 225–235.
-
- Bray N.L. et al. (2016) Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol., 34, 525–527. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

