Onto2Vec: joint vector-based representation of biological entities and their ontology-based annotations
- PMID: 29949999
- PMCID: PMC6022543
- DOI: 10.1093/bioinformatics/bty259
Onto2Vec: joint vector-based representation of biological entities and their ontology-based annotations
Abstract
Motivation: Biological knowledge is widely represented in the form of ontology-based annotations: ontologies describe the phenomena assumed to exist within a domain, and the annotations associate a (kind of) biological entity with a set of phenomena within the domain. The structure and information contained in ontologies and their annotations make them valuable for developing machine learning, data analysis and knowledge extraction algorithms; notably, semantic similarity is widely used to identify relations between biological entities, and ontology-based annotations are frequently used as features in machine learning applications.
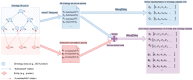
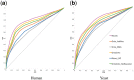
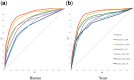
Results: We propose the Onto2Vec method, an approach to learn feature vectors for biological entities based on their annotations to biomedical ontologies. Our method can be applied to a wide range of bioinformatics research problems such as similarity-based prediction of interactions between proteins, classification of interaction types using supervised learning, or clustering. To evaluate Onto2Vec, we use the gene ontology (GO) and jointly produce dense vector representations of proteins, the GO classes to which they are annotated, and the axioms in GO that constrain these classes. First, we demonstrate that Onto2Vec-generated feature vectors can significantly improve prediction of protein-protein interactions in human and yeast. We then illustrate how Onto2Vec representations provide the means for constructing data-driven, trainable semantic similarity measures that can be used to identify particular relations between proteins. Finally, we use an unsupervised clustering approach to identify protein families based on their Enzyme Commission numbers. Our results demonstrate that Onto2Vec can generate high quality feature vectors from biological entities and ontologies. Onto2Vec has the potential to significantly outperform the state-of-the-art in several predictive applications in which ontologies are involved.
Availability and implementation: https://github.com/bio-ontology-research-group/onto2vec.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures

Similar articles
-
Formal axioms in biomedical ontologies improve analysis and interpretation of associated data.Bioinformatics. 2020 Apr 1;36(7):2229-2236. doi: 10.1093/bioinformatics/btz920. Bioinformatics. 2020. PMID: 31821406 Free PMC article.
-
OPA2Vec: combining formal and informal content of biomedical ontologies to improve similarity-based prediction.Bioinformatics. 2019 Jun 1;35(12):2133-2140. doi: 10.1093/bioinformatics/bty933. Bioinformatics. 2019. PMID: 30407490
-
mOWL: Python library for machine learning with biomedical ontologies.Bioinformatics. 2023 Jan 1;39(1):btac811. doi: 10.1093/bioinformatics/btac811. Bioinformatics. 2023. PMID: 36534832 Free PMC article.
-
Semantic similarity in biomedical ontologies.PLoS Comput Biol. 2009 Jul;5(7):e1000443. doi: 10.1371/journal.pcbi.1000443. Epub 2009 Jul 31. PLoS Comput Biol. 2009. PMID: 19649320 Free PMC article. Review.
-
Semantic similarity and machine learning with ontologies.Brief Bioinform. 2021 Jul 20;22(4):bbaa199. doi: 10.1093/bib/bbaa199. Brief Bioinform. 2021. PMID: 33049044 Free PMC article. Review.
Cited by
-
Formal axioms in biomedical ontologies improve analysis and interpretation of associated data.Bioinformatics. 2020 Apr 1;36(7):2229-2236. doi: 10.1093/bioinformatics/btz920. Bioinformatics. 2020. PMID: 31821406 Free PMC article.
-
GeOKG: geometry-aware knowledge graph embedding for Gene Ontology and genes.Bioinformatics. 2025 Mar 29;41(4):btaf160. doi: 10.1093/bioinformatics/btaf160. Bioinformatics. 2025. PMID: 40217132 Free PMC article.
-
KG2Vec: A node2vec-based vectorization model for knowledge graph.PLoS One. 2021 Mar 30;16(3):e0248552. doi: 10.1371/journal.pone.0248552. eCollection 2021. PLoS One. 2021. PMID: 33784319 Free PMC article.
-
Dimensional reduction of phenotypes from 53 000 mouse models reveals a diverse landscape of gene function.Bioinform Adv. 2021 Oct 11;1(1):vbab026. doi: 10.1093/bioadv/vbab026. eCollection 2021. Bioinform Adv. 2021. PMID: 34870209 Free PMC article.
-
Catalyzing Knowledge-Driven Discovery in Environmental Health Sciences through a Community-Driven Harmonized Language.Int J Environ Res Public Health. 2021 Aug 26;18(17):8985. doi: 10.3390/ijerph18178985. Int J Environ Res Public Health. 2021. PMID: 34501574 Free PMC article.
References
-
- Azuaje F. et al. (2005) Ontology-driven similarity approaches to supporting gene functional assessment. In Proceedings of the ISMB’2005 SIG meeting on Bio-ontologies, pp. 9–10.
-
- Bergadano F. (1991) The problem of induction and machine learning. In Proceedings of the 12th International Joint Conference on Artificial Intelligence - Volume 2, IJCAI’91, pp. 1073–1078. Morgan Kaufmann Publishers Inc., San Francisco, CA, USA.
-
- Besold T.R. et al. (2017) Neural-symbolic learning and reasoning: a survey and interpretation. CoRR, abs/1711.03902.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

