Mutational Analysis Identifies Therapeutic Biomarkers in Inflammatory Bowel Disease-Associated Colorectal Cancers
- PMID: 29950348
- PMCID: PMC6193541
- DOI: 10.1158/1078-0432.CCR-17-3713
Mutational Analysis Identifies Therapeutic Biomarkers in Inflammatory Bowel Disease-Associated Colorectal Cancers
Abstract
Purpose: Inflammatory bowel disease-associated colorectal cancers (IBD-CRC) are associated with a higher mortality than sporadic colorectal cancers. The poorly defined molecular pathogenesis of IBD-CRCs limits development of effective prevention, detection, and treatment strategies. We aimed to identify biomarkers using whole-exome sequencing of IBD-CRCs to guide individualized management.Experimental Design: Whole-exome sequencing was performed on 34 formalin-fixed paraffin-embedded primary IBD-CRCs and 31 matched normal lymph nodes. Computational methods were used to identify somatic point mutations, small insertions and deletions, mutational signatures, and somatic copy number alterations. Mismatch repair status was examined.Results: Hypermutation was observed in 27% of IBD-CRCs. All hypermutated cancers were from the proximal colon; all but one of the cancers with hypermutation had defective mismatch repair or somatic mutations in the proofreading domain of DNA POLE Hypermutated IBD-CRCs had increased numbers of predicted neo-epitopes, which could be exploited using immunotherapy. We identified six distinct mutation signatures in IBD-CRCs, three of which corresponded to known mechanisms of mutagenesis. Driver genes were also identified.Conclusions: IBD-CRCs should be evaluated for hypermutation and defective mismatch repair to identify patients with a higher neo-epitope load who may benefit from immunotherapies. Prospective trials are required to determine whether IHC to detect loss of MLH1 expression in dysplastic colonic tissue could identify patients at increased risk of developing IBD-CRC. We identified mutations in genes in IBD-CRCs with hypermutation that might be targeted therapeutically. These approaches would complement and individualize surveillance and treatment programs. Clin Cancer Res; 24(20); 5133-42. ©2018 AACR.
©2018 American Association for Cancer Research.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures

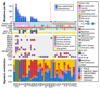



References
-
- Sebastian S, Hernandez V, Myrelid P, Kariv R, Tsianos E, Toruner M, et al. Colorectal cancer in inflammatory bowel disease: results of the 3rd ECCO pathogenesis scientific workshop (I) J Crohns Colitis. 2013;8:5–18. - PubMed
-
- Sanduleanu S, Rutter MD. Interval colorectal cancers in inflammatory bowel disease: the grim statistics and true stories. Gastrointest Endosc Clin N Am. 2014;24:337–348. - PubMed
-
- RCoreTeam. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2015.
-
- Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

