Transmembrane Domains Mediate Intra- and Extracellular Trafficking of Epstein-Barr Virus Latent Membrane Protein 1
- PMID: 29950415
- PMCID: PMC6096814
- DOI: 10.1128/JVI.00280-18
Transmembrane Domains Mediate Intra- and Extracellular Trafficking of Epstein-Barr Virus Latent Membrane Protein 1
Abstract
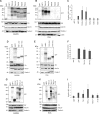
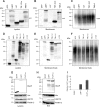
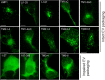
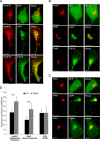
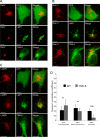
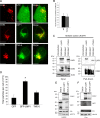
EBV latent membrane protein 1 (LMP1) is released from latently infected tumor cells in small membrane-enclosed extracellular vesicles (EVs). Accumulating evidence suggests that LMP1 is a major driver of EV content and functions. LMP1-modified EVs have been shown to influence recipient cell growth, migration, differentiation, and regulation of immune cell function. Despite the significance of LMP1-modified exosomes, very little is known about how this viral protein enters or manipulates the host EV pathway. In this study, LMP1 deletion mutants were generated to assess protein regions required for EV trafficking. Following transfection of LMP1 or mutant plasmids, EVs were collected by differential centrifugation, and the levels of specific cargo were evaluated by immunoblot analysis. The results demonstrate that, together, the N terminus and transmembrane region 1 of LMP1 are sufficient for efficient sorting into EVs. Consistent with these findings, a mutant lacking the N terminus and transmembrane domains 1 through 4 (TM5-6) failed to be packaged into EVs, and exhibited higher colocalization with endoplasmic reticulum and early endosome markers than the wild-type protein. Surprisingly, TM5-6 maintained the ability to colocalize and form a complex with CD63, an abundant exosome protein that is important for the incorporation of LMP1 into EVs. Other mutations within LMP1 resulted in enhanced levels of secretion, pointing to potential positive and negative regulatory mechanisms for extracellular vesicle sorting of LMP1. These data suggest new functions of the N terminus and transmembrane domains in LMP1 intra- and extracellular trafficking that are likely downstream of an interaction with CD63.IMPORTANCE EBV infection contributes to the development of cancers, such as nasopharyngeal carcinoma, Burkitt lymphoma, Hodgkin's disease, and posttransplant lymphomas, in immunocompromised or genetically susceptible individuals. LMP1 is an important viral protein expressed by EBV in these cancers. LMP1 is secreted in extracellular vesicles (EVs), and the transfer of LMP1-modified EVs to uninfected cells can alter their physiology. Understanding the cellular machinery responsible for sorting LMP1 into EVs is limited, despite the importance of LMP1-modified EVs. Here, we illustrate the roles of different regions of LMP1 in EV packaging. Our results show that the N terminus and TM1 are sufficient to drive LMP1 EV trafficking. We further show the existence of potential positive and negative regulatory mechanisms for LMP1 vesicle sorting. These findings provide a better basis for future investigations to identify the mechanisms of LMP1 targeting to EVs, which could have broad implications in understanding EV cargo sorting.
Keywords: Epstein-Barr virus; exosomes; extracellular vesicles; herpesvirus; microvesicles; protein trafficking; tetraspanin; trafficking.
Copyright © 2018 American Society for Microbiology.
Figures


Similar articles
-
CD63 Regulates Epstein-Barr Virus LMP1 Exosomal Packaging, Enhancement of Vesicle Production, and Noncanonical NF-κB Signaling.J Virol. 2017 Feb 14;91(5):e02251-16. doi: 10.1128/JVI.02251-16. Print 2017 Mar 1. J Virol. 2017. PMID: 27974566 Free PMC article.
-
Tetraspanin CD63 Bridges Autophagic and Endosomal Processes To Regulate Exosomal Secretion and Intracellular Signaling of Epstein-Barr Virus LMP1.J Virol. 2018 Feb 12;92(5):e01969-17. doi: 10.1128/JVI.01969-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29212935 Free PMC article.
-
Epstein-Barr Virus LMP1 Promotes Syntenin-1- and Hrs-Induced Extracellular Vesicle Formation for Its Own Secretion To Increase Cell Proliferation and Migration.mBio. 2020 Jun 16;11(3):e00589-20. doi: 10.1128/mBio.00589-20. mBio. 2020. PMID: 32546618 Free PMC article.
-
Pathogenic role of Epstein-Barr virus latent membrane protein-1 in the development of nasopharyngeal carcinoma.Cancer Lett. 2013 Aug 28;337(1):1-7. doi: 10.1016/j.canlet.2013.05.018. Epub 2013 May 17. Cancer Lett. 2013. PMID: 23689138 Review.
-
Current Trends and Alternative Scenarios in EBV Research.Methods Mol Biol. 2017;1532:1-32. doi: 10.1007/978-1-4939-6655-4_1. Methods Mol Biol. 2017. PMID: 27873264 Review.
Cited by
-
Extracellular Vesicles in the Progression and Therapeutic Resistance of Nasopharyngeal Carcinoma.Cancers (Basel). 2022 May 4;14(9):2289. doi: 10.3390/cancers14092289. Cancers (Basel). 2022. PMID: 35565418 Free PMC article. Review.
-
Extracellular Vesicles in Herpes Viral Spread and Immune Evasion.Front Microbiol. 2018 Oct 25;9:2572. doi: 10.3389/fmicb.2018.02572. eCollection 2018. Front Microbiol. 2018. PMID: 30410480 Free PMC article.
-
Controlling the Revolving and Rotating Motion Direction of Asymmetric Hexameric Nanomotor by Arginine Finger and Channel Chirality.ACS Nano. 2019 Jun 25;13(6):6207-6223. doi: 10.1021/acsnano.8b08849. Epub 2019 May 28. ACS Nano. 2019. PMID: 31067030 Free PMC article. Review.
-
Epstein-Barr virus LMP1 manipulates the content and functions of extracellular vesicles to enhance metastatic potential of recipient cells.PLoS Pathog. 2020 Dec 31;16(12):e1009023. doi: 10.1371/journal.ppat.1009023. eCollection 2020 Dec. PLoS Pathog. 2020. PMID: 33382850 Free PMC article.
-
New insights into Epstein‑Barr virus‑associated tumors: Exosomes (Review).Oncol Rep. 2022 Jan;47(1):13. doi: 10.3892/or.2021.8224. Epub 2021 Nov 15. Oncol Rep. 2022. PMID: 34779497 Free PMC article. Review.
References
-
- Henle G, Henle W, Klein G, Gunven P, Clifford P, Morrow RH, Ziegler JL. 1971. Antibodies to early Epstein-Barr virus-induced antigens in Burkitt's lymphoma. J Natl Cancer Inst 46:861–871. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

