Aedes Anphevirus: an Insect-Specific Virus Distributed Worldwide in Aedes aegypti Mosquitoes That Has Complex Interplays with Wolbachia and Dengue Virus Infection in Cells
- PMID: 29950416
- PMCID: PMC6096813
- DOI: 10.1128/JVI.00224-18
Aedes Anphevirus: an Insect-Specific Virus Distributed Worldwide in Aedes aegypti Mosquitoes That Has Complex Interplays with Wolbachia and Dengue Virus Infection in Cells
Abstract
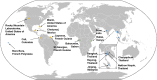
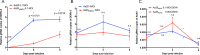
Insect-specific viruses (ISVs) of the yellow fever mosquito Aedes aegypti have been demonstrated to modulate transmission of arboviruses such as dengue virus (DENV) and West Nile virus by the mosquito. The diversity and composition of the virome of A. aegypti, however, remains poorly understood. In this study, we characterized Aedes anphevirus (AeAV), a negative-sense RNA virus from the order Mononegavirales AeAV identified from Aedes cell lines was infectious to both A. aegypti and Aedes albopictus cells but not to three mammalian cell lines. To understand the incidence and genetic diversity of AeAV, we assembled 17 coding-complete and two partial genomes of AeAV from available transcriptome sequencing (RNA-Seq) data. AeAV appears to transmit vertically and be present in laboratory colonies, wild-caught mosquitoes, and cell lines worldwide. Phylogenetic analysis of AeAV strains indicates that as the A. aegypti mosquito has expanded into the Americas and Asia-Pacific, AeAV has evolved into monophyletic African, American, and Asia-Pacific lineages. The endosymbiotic bacterium Wolbachia pipientis restricts positive-sense RNA viruses in A. aegypti Reanalysis of a small RNA library of A. aegypti cells coinfected with AeAV and Wolbachia produces an abundant RNA interference (RNAi) response consistent with persistent virus replication. We found Wolbachia enhances replication of AeAV compared to a tetracycline-cleared cell line, and AeAV modestly reduces DENV replication in vitro The results from our study improve understanding of the diversity and evolution of the virome of A. aegypti and adds to previous evidence that shows Wolbachia does not restrict a range of negative-strand RNA viruses.IMPORTANCE The mosquito Aedes aegypti transmits a number of arthropod-borne viruses (arboviruses), such as dengue virus and Zika virus. Mosquitoes also harbor insect-specific viruses that may affect replication of pathogenic arboviruses in their body. Currently, however, there are only a few insect-specific viruses described from A. aegypti in the literature. Here, we characterize a novel negative-strand virus, AeAV. Meta-analysis of A. aegypti samples showed that it is present in A. aegypti mosquitoes worldwide and is vertically transmitted. Wolbachia-transinfected mosquitoes are currently being used in biocontrol, as they effectively block transmission of several positive-sense RNA viruses in mosquitoes. Our results demonstrate that Wolbachia enhances the replication of AeAV and modestly reduces dengue virus replication in a cell line model. This study expands our understanding of the virome in A. aegypti as well as providing insight into the complexity of the Wolbachia virus restriction phenotype.
Keywords: Aedes aegypti; Mononegavirales; Wolbachia; anphevirus; insect viruses; mosquito; virome.
Copyright © 2018 American Society for Microbiology.
Figures






Similar articles
-
A Novel Anphevirus in Aedes albopictus Mosquitoes Is Distributed Worldwide and Interacts with the Host RNA Interference Pathway.Viruses. 2020 Nov 6;12(11):1264. doi: 10.3390/v12111264. Viruses. 2020. PMID: 33172032 Free PMC article.
-
Cell-Fusing Agent Virus Reduces Arbovirus Dissemination in Aedes aegypti Mosquitoes In Vivo.J Virol. 2019 Aug 28;93(18):e00705-19. doi: 10.1128/JVI.00705-19. Print 2019 Sep 15. J Virol. 2019. PMID: 31243123 Free PMC article.
-
The effects of DENV serotype competition and co-infection on viral kinetics in Wolbachia-infected and uninfected Aedes aegypti mosquitoes.Parasit Vectors. 2021 Jun 9;14(1):314. doi: 10.1186/s13071-021-04816-0. Parasit Vectors. 2021. PMID: 34108021 Free PMC article.
-
Wolbachia-mediated virus blocking in the mosquito vector Aedes aegypti.Curr Opin Insect Sci. 2017 Aug;22:37-44. doi: 10.1016/j.cois.2017.05.005. Epub 2017 May 10. Curr Opin Insect Sci. 2017. PMID: 28805637 Review.
-
Mission Accomplished? We Need a Guide to the 'Post Release' World of Wolbachia for Aedes-borne Disease Control.Trends Parasitol. 2018 Mar;34(3):217-226. doi: 10.1016/j.pt.2017.11.011. Epub 2018 Jan 23. Trends Parasitol. 2018. PMID: 29396201 Review.
Cited by
-
Is the Intergenic Region of Aedes aegypti Totivirus a Recombination Hotspot?Viruses. 2022 Nov 8;14(11):2467. doi: 10.3390/v14112467. Viruses. 2022. PMID: 36366565 Free PMC article.
-
Evolution and diversity of inherited viruses in the Nearctic phantom midge, Chaoborus americanus.Virus Evol. 2022 Mar 10;8(1):veac018. doi: 10.1093/ve/veac018. eCollection 2022. Virus Evol. 2022. PMID: 35356639 Free PMC article.
-
Impact of Hydrogen on the Transcriptome of Sinorhizobium meliloti 1021 Using RNA-sequencing Technology.Pol J Microbiol. 2020;69(1):1-10. doi: 10.33073/pjm-2020-006. Pol J Microbiol. 2020. PMID: 32067442 Free PMC article.
-
Virome profiling of fig wasps (Ceratosolen spp.) reveals virus diversity spanning four realms.Virology. 2024 Mar;591:109992. doi: 10.1016/j.virol.2024.109992. Epub 2024 Jan 10. Virology. 2024. PMID: 38246037 Free PMC article.
-
Bunyamwera Virus Infection of Wolbachia-Carrying Aedes aegypti Mosquitoes Reduces Wolbachia Density.Viruses. 2024 Aug 21;16(8):1336. doi: 10.3390/v16081336. Viruses. 2024. PMID: 39205310 Free PMC article.
References
-
- Shriram AN, Sivan A, Sugunan AP. 2017. Spatial distribution of Aedes aegypti and Aedes albopictus in relation to geo-ecological features in South Andaman, Andaman and Nicobar Islands, India. Bull Entomol Res 3:166–174. - PubMed
-
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi: 10.1038/nature12060. - DOI - PMC - PubMed
-
- European Centre for Disease Prevention and Control. 2015. Rapid risk assessment: Zika virus epidemic in the Americas: potential association with microcephaly and Guillain-Barré syndrome. European Centre for Disease Prevention and Control, Solna, Sweden: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publicat....
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

