Tomato Yellow Leaf Curl Virus V2 Interacts with Host Histone Deacetylase 6 To Suppress Methylation-Mediated Transcriptional Gene Silencing in Plants
- PMID: 29950418
- PMCID: PMC6146709
- DOI: 10.1128/JVI.00036-18
Tomato Yellow Leaf Curl Virus V2 Interacts with Host Histone Deacetylase 6 To Suppress Methylation-Mediated Transcriptional Gene Silencing in Plants
Abstract
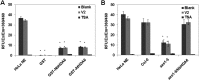
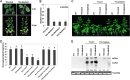
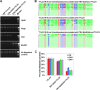
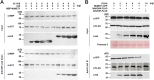
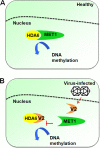
Cytosine DNA methylation is a conserved epigenetic silencing mechanism that defends against biotic stresses such as geminivirus infection. As a countermeasure, geminiviruses encode proteins that inhibit methylation and transcriptional gene silencing (TGS). Previous studies showed that V2 protein of Tomato yellow leaf curl virus (TYLCV) functions as a TGS suppressor. However, how V2 mediates TGS suppression remains unknown. Here we show that V2 interacts directly with a Nicotiana benthamiana histone deacetylase 6 (NbHDA6), a homolog of Arabidopsis HDA6 (AtHDA6), known to be involved in gene silencing in cooperation with methyltransferase 1 (MET1). NbHDA6 genetically complemented a late-flowering phenotype and restored histone deacetylation of an AtHDA6 mutant. Furthermore, our investigation showed that NbHDA6 displayed histone deacetylase enzymatic activity, which was not inhibited by V2. Genetic analysis revealed that silencing of NbHDA6 expression resulted in enhanced susceptibility to TYLCV infection. In addition, methylation-sensitive PCR and bisulfite sequencing analysis showed that silencing of NbHDA6 expression caused reduced DNA methylation of the viral genome in infected plants. HDA6 was previously shown to recruit and physically interact with MET1 to function in gene silencing. Using competitive pulldown and coimmunoprecipitation assays, we demonstrated that V2 did not interact but competed with NbMET1 for direct binding to NbHDA6. These findings suggest that V2 interacts with host HDA6 and interferes with the recruitment of MET1 by HDA6, resulting in decreased methylation of the viral DNA genome by TGS with a concomitant increase in host susceptibility to TYLCV infection.IMPORTANCE Plants employ repressive viral genome methylation as an epigenetic defense against geminiviruses. In turn, geminiviruses encode proteins that inhibit methylation by TGS. Previous studies showed that TYLCV V2 can efficiently suppress TGS, but the mechanism remains unknown. We showed that V2 interacted with NbHDA6 but did not inhibit its enzymatic activity. As HDA6 is known to be involved in gene silencing in cooperation with MET1, we explored the relationship between V2, NbMET1, and NbHDA6. Our investigation showed that V2 did not interact but competed with NbMET1 for direct binding to NbHDA6. To our knowledge, this is the first report that viral proteins inhibit TGS by interacting with histone deacetylase but not by blocking the methyl cycle. This work provides an additional mechanism for TGS suppression by geminiviruses.
Keywords: DNA methylation; MET1; Tomato yellow leaf curl virus; V2; geminivirus; histone deacetylase 6 (HDA6); methyltransferase 1; suppressor; suppressor of RNA silencing; transcriptional gene silencing (TGS).
Copyright © 2018 American Society for Microbiology.
Figures



References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

