Geminivirus Replication Protein Impairs SUMO Conjugation of Proliferating Cellular Nuclear Antigen at Two Acceptor Sites
- PMID: 29950424
- PMCID: PMC6146701
- DOI: 10.1128/JVI.00611-18
Geminivirus Replication Protein Impairs SUMO Conjugation of Proliferating Cellular Nuclear Antigen at Two Acceptor Sites
Abstract
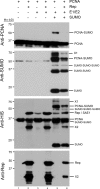
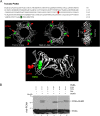
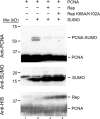
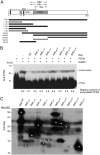
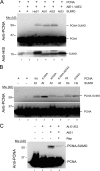
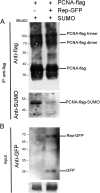
Geminiviruses are DNA viruses that replicate in nuclei of infected plant cells using the plant DNA replication machinery, including PCNA (proliferating cellular nuclear antigen), a cofactor that orchestrates genome duplication and maintenance by recruiting crucial players to replication forks. These viruses encode a multifunctional protein, Rep, which is essential for viral replication, induces the accumulation of the host replication machinery, and interacts with several host proteins, including PCNA and the SUMO E2 conjugation enzyme (SCE1). Posttranslational modification of PCNA by ubiquitin or SUMO plays an essential role in the switching of PCNA between interacting partners during DNA metabolism processes (e.g., replication, recombination, and repair, etc.). In yeast, PCNA sumoylation has been associated with DNA repair involving homologous recombination (HR). Previously, we reported that ectopic Rep expression results in very specific changes in the sumoylation pattern of plant cells. In this work, we show, using a reconstituted sumoylation system in Escherichia coli, that tomato PCNA is sumoylated at two residues, K254 and K164, and that coexpression of the geminivirus protein Rep suppresses sumoylation at these lysines. Finally, we confirm that PCNA is sumoylated in planta and that Rep also interferes with PCNA sumoylation in plant cells.IMPORTANCE SUMO adducts have a key role in regulating the activity of animal and yeast PCNA on DNA repair and replication. Our work demonstrates for the first time that sumoylation of plant PCNA occurs in plant cells and that a plant virus interferes with this modification. This work marks the importance of sumoylation in allowing viral infection and replication in plants. Moreover, it constitutes a prime example of how viral proteins interfere with posttranslational modifications of selected host factors to create a proper environment for infection.
Keywords: PCNA; Rep; SUMO; begomovirus; geminivirus; homologous recombination; sumoylation.
Copyright © 2018 American Society for Microbiology.
Figures

References
-
- Jeske H. 2009. Geminiviruses. Curr Top Microbiol Immunol 331:185–226. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

