Exploring the links between cancer and placenta development
- PMID: 29950452
- PMCID: PMC6030113
- DOI: 10.1098/rsob.180081
Exploring the links between cancer and placenta development
Abstract
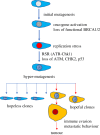
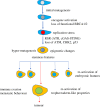
The development of metastatic cancer is a multistage process, which often requires decades to complete. Impairments in DNA damage control and DNA repair in cancer cell precursors generate genetically heterogeneous cell populations. However, despite heterogeneity most solid cancers have stereotypical behaviours, including invasiveness and suppression of immune responses that can be unleashed with immunotherapy targeting lymphocyte checkpoints. The mechanisms leading to the acquisition of stereotypical properties remain poorly understood. Reactivation of embryonic development processes in cells with unstable genomes might contribute to tumour expansion and metastasis formation. However, it is unclear whether these events are linked to immune response modulation. Tumours and embryos have non-self-components and need to avoid immune responses in their microenvironment. In mammalian embryos, neo-antigens are of paternal origin, while in tumour cells DNA mismatch repair and replication defects generate them. Inactivation of the maternal immune response towards the embryo, which occurs at the placental-maternal interface, is key to ensuring embryonic development. This regulation is accomplished by the trophoblast, which mimics several malignant cell features, including the ability to invade normal tissues and to avoid host immune responses, often adopting the same cancer immunoediting strategies. A better understanding as to whether and how genotoxic stress promotes cancer development through reactivation of programmes occurring during early stages of mammalian placentation could help to clarify resistance to drugs targeting immune checkpoint and DNA damage responses and to develop new therapeutic strategies to eradicate cancer.
Keywords: DNA damage response; DNA repair; cancer.
© 2018 The Authors.
Conflict of interest statement
We declare we have no competing interests.
Figures
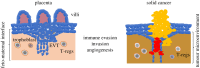
References
-
- Burrell RA, McGranahan N, Bartek J, Swanton C. 2013. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501, 338–345. (doi:10.1038/nature12625) - DOI - PubMed
-
- Patel AP, et al. 2014. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401. (doi:10.1126/science.1254257) - DOI - PMC - PubMed
-
- Torres CM, et al. 2016. The linker histone H1.0 generates epigenetic and functional intratumor heterogeneity. Science 353, aaf1644 (doi:10.1126/science.aaf1644) - DOI - PMC - PubMed
-
- Zeman MK, Cimprich KA. 2014. Causes and consequences of replication stress. Nat. Cell Biol. 16, 2–9. (doi:10.1038/ncb2897) - DOI - PMC - PubMed
-
- Taylor EM, Lindsay HD. 2016. DNA replication stress and cancer: cause or cure? Future Oncol. 12, 221–237. (doi:10.2217/fon.15.292) - DOI - PubMed

