C-NHEJ without indels is robust and requires synergistic function of distinct XLF domains
- PMID: 29950655
- PMCID: PMC6021437
- DOI: 10.1038/s41467-018-04867-5
C-NHEJ without indels is robust and requires synergistic function of distinct XLF domains
Abstract
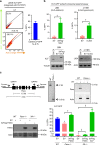
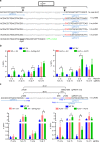
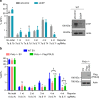
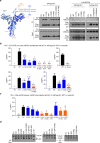
To investigate the fidelity of canonical non-homologous end joining (C-NHEJ), we developed an assay to detect EJ between distal ends of two Cas9-induced chromosomal breaks that are joined without causing insertion/deletion mutations (indels). Here we find that such EJ requires several core C-NHEJ factors, including XLF. Using variants of this assay, we find that C-NHEJ is required for EJ events that use 1-2, but not ≥3, nucleotides of terminal microhomology. We also investigated XLF residues required for EJ without indels, finding that one of two binding domains is essential (L115 or C-terminal lysines that bind XRCC4 and KU/DNA, respectively), and that disruption of one of these domains sensitizes XLF to mutations that affect its dimer interface, which we examined with molecular dynamic simulations. Thus, C-NHEJ, including synergistic function of distinct XLF domains, is required for EJ of chromosomal breaks without indels.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

