Complexes formed by mutant p53 and their roles in breast cancer
- PMID: 29950894
- PMCID: PMC6011883
- DOI: 10.2147/BCTT.S145826
Complexes formed by mutant p53 and their roles in breast cancer
Abstract
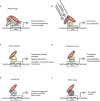
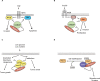
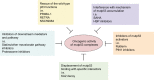
Breast cancer is the most frequently diagnosed malignancy in women, and mutations in the tumor suppressor p53 are commonly detected in the most aggressive subtypes. The majority of TP53 gene alterations are missense substitutions, leading to expression of mutant forms of the p53 protein that are frequently detected at high levels in cancer cells. P53 mutants not only lose the physiological tumor-suppressive activity of the wild-type p53 protein but also acquire novel powerful oncogenic functions, referred to as gain of function, that may actively confer a selective advantage during tumor progression. Some of the best-characterized oncogenic activities of mutant p53 are mediated by its ability to form aberrant protein complexes with other transcription factors or proteins not directly related to gene transcription. The set of cellular proteins available to interact with mutant p53 is dependent on cell type and extensively affected by environmental signals, so the prognostic impact of p53 mutation is complex. Specific functional interactions of mutant p53 can profoundly impact homeostasis of breast cancer cells, reprogramming gene expression in response to specific extracellular inputs or cell-intrinsic conditions. The list of protein complexes involving mutant p53 in breast cancer is continuously growing, as is the number of oncogenic phenotypes in which they could be involved. In consideration of the functional impact of such complexes, key interactions of mutant p53 may be exploited as potential targets for development of therapies aimed at defusing the oncogenic potential of p53 mutation.
Keywords: cancer-cell homeostasis; mutant p53 gain of function; protein–protein interactions; targeted therapy.
Conflict of interest statement
Disclosure The authors report no conflicts of interest in this work.
Figures
Similar articles
-
Mutant p53 in Cancer Progression and Targeted Therapies.Front Oncol. 2020 Nov 6;10:595187. doi: 10.3389/fonc.2020.595187. eCollection 2020. Front Oncol. 2020. PMID: 33240819 Free PMC article. Review.
-
Mutant p53 accumulation in human breast cancer is not an intrinsic property or dependent on structural or functional disruption but is regulated by exogenous stress and receptor status.J Pathol. 2014 Jul;233(3):238-46. doi: 10.1002/path.4356. Epub 2014 May 21. J Pathol. 2014. PMID: 24687952
-
Targeting mutant p53: a key player in breast cancer pathogenesis and beyond.Cell Commun Signal. 2024 Oct 10;22(1):484. doi: 10.1186/s12964-024-01863-9. Cell Commun Signal. 2024. PMID: 39390510 Free PMC article. Review.
-
Transcriptional Regulation by Wild-Type and Cancer-Related Mutant Forms of p53.Cold Spring Harb Perspect Med. 2017 Feb 1;7(2):a026054. doi: 10.1101/cshperspect.a026054. Cold Spring Harb Perspect Med. 2017. PMID: 27836911 Free PMC article. Review.
-
The rebel angel: mutant p53 as the driving oncogene in breast cancer.Carcinogenesis. 2012 Nov;33(11):2007-17. doi: 10.1093/carcin/bgs232. Epub 2012 Jul 20. Carcinogenesis. 2012. PMID: 22822097 Free PMC article. Review.
Cited by
-
Prognostic Value of Intracellular Transcription of Factors HIF-1α and p53 and Their Relation to Estradiol and TNM Parameters of Breast Cancer Tissues in Women with Invasive Ductal Carcinoma in Thi-Qar Province, Iraq.Arch Razi Inst. 2022 Aug 31;77(4):1341-1348. doi: 10.22092/ARI.2022.357640.2080. eCollection 2022 Aug. Arch Razi Inst. 2022. PMID: 36883155 Free PMC article.
-
FBXO2 promotes the progression of papillary thyroid carcinoma through the p53 pathway.Sci Rep. 2024 Sep 29;14(1):22574. doi: 10.1038/s41598-024-73455-z. Sci Rep. 2024. PMID: 39343799 Free PMC article.
-
Detection of gene mutations in gastric cancer tissues using a commercial sequencing panel.Mol Clin Oncol. 2019 Nov;11(5):455-460. doi: 10.3892/mco.2019.1926. Epub 2019 Sep 24. Mol Clin Oncol. 2019. PMID: 31620276 Free PMC article.
-
Structural studies of antitumor compounds that target the RING domain of MDM2.Protein Sci. 2022 Aug;31(8):e4367. doi: 10.1002/pro.4367. Protein Sci. 2022. Retraction in: Protein Sci. 2023 Jun;32(6):e4657. doi: 10.1002/pro.4657. PMID: 35900024 Free PMC article. Retracted.
-
Identification of potential target genes of breast cancer in response to Chidamide treatment.Front Mol Biosci. 2022 Nov 8;9:999582. doi: 10.3389/fmolb.2022.999582. eCollection 2022. Front Mol Biosci. 2022. PMID: 36425653 Free PMC article.
References
-
- Dumay A, Feugeas JP, Wittmer E, et al. Distinct tumor protein p53 mutants in breast cancer subgroups. Int J Cancer. 2013;132(5):1227–1231. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

