New operational taxonomic units of Enterocytozoon in three marsupial species
- PMID: 29954462
- PMCID: PMC6022301
- DOI: 10.1186/s13071-018-2954-x
New operational taxonomic units of Enterocytozoon in three marsupial species
Abstract
Background: Enterocytozoon bieneusi is a microsporidian, commonly found in animals, including humans, in various countries. However, there is scant information about this microorganism in Australasia. In the present study, we conducted the first molecular epidemiological investigation of E. bieneusi in three species of marsupials (Macropus giganteus, Vombatus ursinus and Wallabia bicolor) living in the catchment regions which supply the city of Melbourne with drinking water.
Methods: Genomic DNAs were extracted from 1365 individual faecal deposits from these marsupials, including common wombat (n = 315), eastern grey kangaroo (n = 647) and swamp wallaby (n = 403) from 11 catchment areas, and then individually tested using a nested PCR-based sequencing approach employing the internal transcribed spacer (ITS) and small subunit (SSU) of nuclear ribosomal DNA as genetic markers.
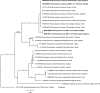
Results: Enterocytozoon bieneusi was detected in 19 of the 1365 faecal samples (1.39%) from wombat (n = 1), kangaroos (n = 13) and wallabies (n = 5). The analysis of ITS sequence data revealed a known (designated NCF2) and four new (MWC_m1 to MWC_m4) genotypes of E. bieneusi. Phylogenetic analysis of ITS sequence data sets showed that MWC_m1 (from wombat) clustered with NCF2, whereas genotypes MWC_m2 (kangaroo and wallaby), MWC_m3 (wallaby) and MWC_m4 (kangaroo) formed a new, divergent clade. Phylogenetic analysis of SSU sequence data revealed that genotypes MWC_m3 and MWC_m4 formed a clade that was distinct from E. bieneusi. The genetic distinctiveness of these two genotypes suggests that they represent a new species of Enterocytozoon.
Conclusions: Further investigations of Enterocytozoon spp. from macropods and other animals will assist in clarifying the taxonomy and epidemiology of these species in Australia and elsewhere, and in assessing the public health risk of enterocytozoonosis.
Keywords: Australia; Common wombat; Eastern grey kangaroo; Enterocytozoon bieneusi; Genotypes; Operational taxonomic units; Prevalence; Swamp wallaby.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
-
- Molina JM, Sarfati C, Beauvais B, Lémann M, Lesourd A, Ferchal F, et al. Intestinal microsporidiosis in human immunodeficiency virus-infected patients with chronic unexplained diarrhea: prevalence and clinical and biologic features. J Infect Dis. 1993;167:217–221. doi: 10.1093/infdis/167.1.217. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

