Comprehensive ADP-ribosylome analysis identifies tyrosine as an ADP-ribose acceptor site
- PMID: 29954836
- PMCID: PMC6073207
- DOI: 10.15252/embr.201745310
Comprehensive ADP-ribosylome analysis identifies tyrosine as an ADP-ribose acceptor site
Abstract
Despite recent mass spectrometry (MS)-based breakthroughs, comprehensive ADP-ribose (ADPr)-acceptor amino acid identification and ADPr-site localization remain challenging. Here, we report the establishment of an unbiased, multistep ADP-ribosylome data analysis workflow that led to the identification of tyrosine as a novel ARTD1/PARP1-dependent in vivo ADPr-acceptor amino acid. MS analyses of in vitro ADP-ribosylated proteins confirmed tyrosine as an ADPr-acceptor amino acid in RPS3A (Y155) and HPF1 (Y238) and demonstrated that trans-modification of RPS3A is dependent on HPF1. We provide an ADPr-site Localization Spectra Database (ADPr-LSD), which contains 288 high-quality ADPr-modified peptide spectra, to serve as ADPr spectral references for correct ADPr-site localizations.
Keywords: ADP‐ribosylation; ARTD1/PARP1; HPF1; genotoxic stress; tyrosine ADP‐ribosylation.
© 2018 The Authors.
Figures
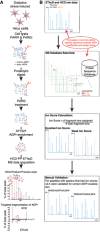
Schematic of the sample preparation and HCD‐PP‐EThcD/HCD MS acquisition method workflow employed by Bilan et al 4 to generate the EThcD and HCD datasets used here.
Schematic of the bioinformatics workflow employed here. In the initial phase of these analyses, variable ADPr PTM search parameters were opened to include all 20 amino acids as potential ADPr acceptors. Ion scores were then calculated to help identify the best‐quality ADPr‐site localizations within modified peptides and peptides for manual validation of ADPr‐site localization. Based on these manual validations, the search parameters were refined to include S, D, E, R, K, and Y (SDERKY) or S, R, and Y (SRY) as ADPr‐acceptor amino acids and the workflow was re‐applied.
The ADPr‐site localization results from the initial (20 amino acids) searches and refined searches were compared. The flowchart depicts how ADPr‐acceptor amino acid identifications changed for the 181 EThcD ADPr localization training spectra as the variable ADPr search parameters were refined. Importantly, these analyses identified novel tyrosine (Y) ADPr modifications at each phase of our analyses.
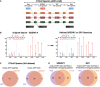
The left panel depicts an EThcD fragmentation spectrum of an ARTD1 peptide in which the original search localized the ADPr modification to A5 of the peptide. The right panel is the same EThcD spectrum annotated after the data were searched using the refined ADPr search parameters developed here. The spectrum annotations on the right provide stronger evidence that this ADPr modification is on S3.
Comparison of total number of unique ADP‐ribosylated peptides and unique ADPr sites identified (Mascot localization probability > 60%) in the SDERKY and SRY searches.
Comparison of the ADPr‐site localizations (Mascot localization probability > 60%) for ADP‐ribosylated peptides that were identified by both EThcD and HCD fragmentation methods.
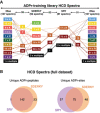
ADPr‐site localization results from the initial (20 amino acids) searches and refined searches were compared. The flowchart depicts how ADPr‐acceptor amino acid localizations changed for the 107 HCD ADPr localization training spectra as the variable ADPr search parameters were refined.
Comparison of total number of unique ADP‐ribosylated peptides and unique ADPr‐sites identified (Mascot localization probability > 60%) from HCD spectra in the SDERKY and SRY searches.
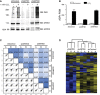
α‐ARTD1, α‐ARTD2, and α‐PAR Western blot analyses of untreated and H2O2‐treated shControl, shARTD1, and shARTD2 HeLa cells lysates confirm specific knockdown of ARTD1 or ARTD2 and specific inhibition of H2O2‐induced PAR following loss of ARTD1.
Number of ADPr‐PSMs identified in untreated and H2O2‐treated HeLa cells following ARTD1 or ARTD2 knockdown. Error bars represent standard deviations of the mean (n = 3).
Heat map of Spearman's rank correlation coefficient and multi‐scatter plots showing reproducibility between cell line ADP‐ribosylomes.
Heat map of unsupervised clustering of the z‐score of ADPr‐site abundance in the different cell lines.
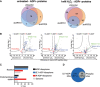
Venn diagrams indicate the overlap of ADP‐ribosylated proteins identified in untreated and H2O2‐treated HeLa cells following ARTD1 or ARTD2 knockdown.
Volcano plots comparing ADP‐ribosylated peptides identified in the different cell lines and conditions. ADP‐ribosylation sites confirmed by EThcD spectra are color‐coded for S‐ADPr (blue), Y‐ADPr (green), and R‐ADPr (red) modifications. Serine (DDX21), tyrosine (RPS3A), and arginine (PDIA3) modifications that were identified within our ADPr localization training spectra library are annotated. The black hyperbolic line represents a permutation‐based false discovery rate (FDR) of 5% and a minimal fold change of 2.
Gene ontology cellular localization annotation of ADP‐ribosylated proteins following H2O2 treatment: Only SRY ADP‐ribosylated proteins with a Mascot localization score > 90% were considered.
Comparison of the S‐ and Y‐ADP‐ribosylation identified here with the HeLa phosphoproteome identified by Sharma et al 20.
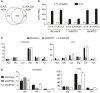
- A
Venn diagram depicts the overlap of the total number of unique ADPr‐peptides identified following H2O2 treatment of shControl, shARTD1, and shARTD2 cells by Mascot‐based searches that considered either S, R, and Y or S, D, E, R, K, and Y as variable ADPr‐acceptor amino acids. Note that ADPr‐site localization within the peptide was ignored for this analysis.
- B
Total number of ADPr‐peptide spectrum matches (PSMs) identified in the different cell lines and treatment conditions for SRY and SDERKY searches. Error bars represent standard deviations of the mean (n = 3).
- C, D
Total number of ADPr‐sites localized (Mascot localization score > 60%) in the SDERKY (c) and SRY (d) searches. Error bars represent standard deviations of the mean (n = 3).
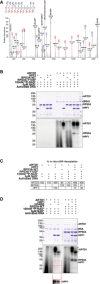
High‐resolution EThcD fragmentation spectrum of an RPS3A peptide modified with ADP‐ribose on Y155.
Recombinant RPS3A‐WT and RPS3A‐Y155F were in vitro ADP‐ribosylated with [32P]‐NAD+, activated DNA, and recombinant ARTD1 in the presence or absence of recombinant HPF1. Samples were resolved by SDS–PAGE and analyzed by autoradiography. Top: Coomassie blue‐stained polyacrylamide gel; bottom: autoradiograph with levels adjusted to enhance exposure.
Relative percentages of the in vitro ADP‐ribosylation (radioactive signal) of ARTD1 (relative to radioactive signal with [32P]‐NAD+ and activated DNA alone), RPS3A‐WT, and RPS3A‐Y155F (relative to radioactive signal for RPS3A‐WT for each condition) are shown (n = 3).
Recombinant ARTD1 and RPS3A‐WT were in vitro ADP‐ribosylated with [32P]‐NAD+ and activated DNA in the presence or absence of recombinant HPF1. The indicated samples were then treated with recombinant human PARG (hPARG) to reduce PAR modifications to MAR modifications. The samples were resolved by SDS–PAGE and analyzed by autoradiography. Top: Coomassie blue‐stained polyacrylamide gel; middle: autoradiograph with levels adjusted to enhance exposure; bottom inset: autoradiograph of HPF1 section only with enhanced exposure.
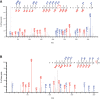
High‐resolution EThcD fragmentation spectra of the HPF1 peptide TFHGAGLVVPVDKNDVGYRELPETDADLKRELPETDADLKR identified during re‐analysis of in vivo HeLa H2O2‐treated MS datasets. This spectrum supports ADP‐ribosylation of Y238.
High‐resolution EThcD fragmentation spectra of HPF1 peptide TFHGAGLVVPVDKNDVGYRELPETDADLKR acquired during MS analysis of ARTD1/HPF1 in vitro ADP‐ribosylation assays. This spectrum also supports ADP‐ribosylation of Y238.
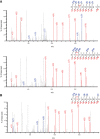
High‐resolution EThcD fragmentation spectra of RPS3A peptide KTSYAQHQQVR acquired during MS analysis of ARTD1/HPF1/RPS3A‐WT in vitro ADP‐ribosylation assays. Top spectrum supports ADPr localization on Y155, and the bottom spectrum supports ADPr localization on S154.
High‐resolution EThcD fragmentation spectrum of RPS3A peptide KTSFAQHQQVR acquired during MS analysis of ARTD1/HPF1/RPS3A‐Y155F in vitro ADP‐ribosylation assays. Spectrum supports ADPr localization on S154.
References
-
- Barkauskaite E, Jankevicius G, Ahel I (2015) Structures and mechanisms of enzymes employed in the synthesis and degradation of PARP‐dependent protein ADP‐ribosylation. Mol Cell 58: 935–946 - PubMed
-
- Hottiger MO (2015) Nuclear ADP‐ribosylation and its role in chromatin plasticity, cell differentiation, and epigenetics. Annu Rev Biochem 84: 227–263 - PubMed
-
- Bilan V, Leutert M, Nanni P, Panse C, Hottiger MO (2017) Combining HCD and EThcD fragmentation in a product dependent‐manner confidently assigns proteome‐wide ADP‐ribose acceptor sites. Anal Chem 89: 1523–1530 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

