Methanotrophy across a natural permafrost thaw environment
- PMID: 29955139
- PMCID: PMC6155033
- DOI: 10.1038/s41396-018-0065-5
Methanotrophy across a natural permafrost thaw environment
Abstract
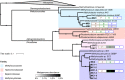
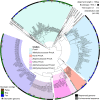
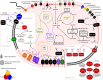
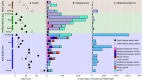
The fate of carbon sequestered in permafrost is a key concern for future global warming as this large carbon stock is rapidly becoming a net methane source due to widespread thaw. Methane release from permafrost is moderated by methanotrophs, which oxidise 20-60% of this methane before emission to the atmosphere. Despite the importance of methanotrophs to carbon cycling, these microorganisms are under-characterised and have not been studied across a natural permafrost thaw gradient. Here, we examine methanotroph communities from the active layer of a permafrost thaw gradient in Stordalen Mire (Abisko, Sweden) spanning three years, analysing 188 metagenomes and 24 metatranscriptomes paired with in situ biogeochemical data. Methanotroph community composition and activity varied significantly as thaw progressed from intact permafrost palsa, to partially thawed bog and fully thawed fen. Thirteen methanotroph population genomes were recovered, including two novel genomes belonging to the uncultivated upland soil cluster alpha (USCα) group and a novel potentially methanotrophic Hyphomicrobiaceae. Combined analysis of porewater δ13C-CH4 isotopes and methanotroph abundances showed methane oxidation was greatest below the oxic-anoxic interface in the bog. These results detail the direct effect of thaw on autochthonous methanotroph communities, and their consequent changes in population structure, activity and methane moderation potential.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Tarnocai C, Riley WJ, Canadell JG, Schuur EAG, Kuhry P, Mazhitova G, Zimov S. Soil organic carbon pools in the northern circumpolar permafrost region. Glob Biogeochem Cycles. 2009;23:GB2023. doi: 10.1029/2008GB003327. - DOI
-
- Koven CD, Riley WJ, Stern A. Analysis of permafrost thermal dynamics and response to climate change in the CMIP5 earth system models. J Clim. 2013;26:1877–1900. doi: 10.1175/JCLI-D-12-00228.1. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

