Genome-scale analysis identifies paralog lethality as a vulnerability of chromosome 1p loss in cancer
- PMID: 29955178
- PMCID: PMC6143899
- DOI: 10.1038/s41588-018-0155-3
Genome-scale analysis identifies paralog lethality as a vulnerability of chromosome 1p loss in cancer
Abstract
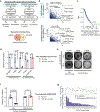
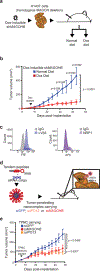
Functional redundancy shared by paralog genes may afford protection against genetic perturbations, but it can also result in genetic vulnerabilities due to mutual interdependency1-5. Here, we surveyed genome-scale short hairpin RNA and CRISPR screening data on hundreds of cancer cell lines and identified MAGOH and MAGOHB, core members of the splicing-dependent exon junction complex, as top-ranked paralog dependencies6-8. MAGOHB is the top gene dependency in cells with hemizygous MAGOH deletion, a pervasive genetic event that frequently occurs due to chromosome 1p loss. Inhibition of MAGOHB in a MAGOH-deleted context compromises viability by globally perturbing alternative splicing and RNA surveillance. Dependency on IPO13, an importin-β receptor that mediates nuclear import of the MAGOH/B-Y14 heterodimer9, is highly correlated with dependency on both MAGOH and MAGOHB. Both MAGOHB and IPO13 represent dependencies in murine xenografts with hemizygous MAGOH deletion. Our results identify MAGOH and MAGOHB as reciprocal paralog dependencies across cancer types and suggest a rationale for targeting the MAGOHB-IPO13 axis in cancers with chromosome 1p deletion.
Figures


Similar articles
-
Multifaceted roles of MAGOH Proteins.Mol Biol Rep. 2023 Feb;50(2):1931-1941. doi: 10.1007/s11033-022-07904-1. Epub 2022 Nov 17. Mol Biol Rep. 2023. PMID: 36396768 Review.
-
The paralogues MAGOH and MAGOHB are oncogenic factors in high-grade gliomas and safeguard the splicing of cell division and cell cycle genes.RNA Biol. 2023 Jan;20(1):311-322. doi: 10.1080/15476286.2023.2221511. RNA Biol. 2023. PMID: 37294214 Free PMC article.
-
Two mammalian MAGOH genes contribute to exon junction complex composition and nonsense-mediated decay.RNA Biol. 2013 Aug;10(8):1291-8. doi: 10.4161/rna.25827. Epub 2013 Jul 23. RNA Biol. 2013. PMID: 23917022 Free PMC article.
-
MAGOH and MAGOHB Knockdown in Melanoma Cells Decreases Nonsense-Mediated Decay Activity and Promotes Apoptosis via Upregulation of GADD45A.Cells. 2022 Nov 30;11(23):3859. doi: 10.3390/cells11233859. Cells. 2022. PMID: 36497117 Free PMC article.
-
Diverse Roles of the Exon Junction Complex Factors in the Cell Cycle, Cancer, and Neurodevelopmental Disorders-Potential for Therapeutic Targeting.Int J Mol Sci. 2022 Sep 8;23(18):10375. doi: 10.3390/ijms231810375. Int J Mol Sci. 2022. PMID: 36142288 Free PMC article. Review.
Cited by
-
Minor intron-containing genes as an ancient backbone for viral infection?PNAS Nexus. 2024 Jan 18;3(1):pgad479. doi: 10.1093/pnasnexus/pgad479. eCollection 2024 Jan. PNAS Nexus. 2024. PMID: 38274120 Free PMC article.
-
A Division of Labor between YAP and TAZ in Non-Small Cell Lung Cancer.Cancer Res. 2020 Oct 1;80(19):4145-4157. doi: 10.1158/0008-5472.CAN-20-0125. Epub 2020 Aug 14. Cancer Res. 2020. PMID: 32816858 Free PMC article.
-
Aneuploidy as a driver of human cancer.Nat Genet. 2024 Oct;56(10):2014-2026. doi: 10.1038/s41588-024-01916-2. Epub 2024 Oct 2. Nat Genet. 2024. PMID: 39358600 Review.
-
Patterns of Aneuploidy and Signaling Consequences in Cancer.Cancer Res. 2024 Aug 15;84(16):2575-2587. doi: 10.1158/0008-5472.CAN-24-0169. Cancer Res. 2024. PMID: 38924459 Free PMC article. Review.
-
Genoppi is an open-source software for robust and standardized integration of proteomic and genetic data.Nat Commun. 2021 May 10;12(1):2580. doi: 10.1038/s41467-021-22648-5. Nat Commun. 2021. PMID: 33972534 Free PMC article.
References
REFERENCES (FOR MAIN TEXT)
-
- Diss G et al. Gene duplication can impart fragility, not robustness, in the yeast protein interaction network. Science 355, 630–634 (2017). - PubMed
-
- Frei E Gene deletion: a new target for cancer chemotherapy. The Lancet 342, 662–664 (1993). - PubMed
-
- McDonald ER et al. Project DRIVE: A Compendium of Cancer Dependencies and Synthetic Lethal Relationships Uncovered by Large-Scale, Deep RNAi Screening. Cell 170, 577–592.e10 (2017). - PubMed
REFERENCES (METHODS ONLY)
-
- Lippa MS et al. Expression of anti-apoptotic factors modulates Apo2L/TRAIL resistance in colon carcinoma cells. Apoptosis Int. J. Program. Cell Death 12, 1465–1478 (2007). - PubMed
-
- Brown CY et al. Robust, reversible gene knockdown using a single lentiviral short hairpin RNA vector. Hum. Gene Ther. 21, 1005–1017 (2010). - PubMed
-
- Root DE, Hacohen N, Hahn WC, Lander ES & Sabatini DM Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat. Methods 3, 715–719 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

