Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation
- PMID: 29955203
- PMCID: PMC6020082
- DOI: 10.1016/j.simyco.2018.05.001
Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation
Abstract
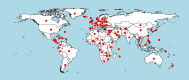
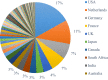
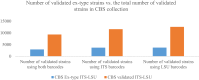
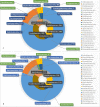
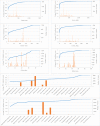
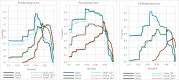
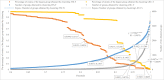
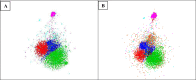
Species identification lies at the heart of biodiversity studies that has in recent years favoured DNA-based approaches. Microbial Biological Resource Centres are a rich source for diverse and high-quality reference materials in microbiology, and yet the strains preserved in these biobanks have been exploited only on a limited scale to generate DNA barcodes. As part of a project funded in the Netherlands to barcode specimens of major national biobanks, sequences of two nuclear ribosomal genetic markers, the Internal Transcribed Spaces and 5.8S gene (ITS) and the D1/D2 domain of the 26S Large Subunit (LSU), were generated as DNA barcode data for ca. 100 000 fungal strains originally assigned to ca. 17 000 species in the CBS fungal biobank maintained at the Westerdijk Fungal Biodiversity Institute, Utrecht. Using more than 24 000 DNA barcode sequences of 12 000 ex-type and manually validated filamentous fungal strains of 7 300 accepted species, the optimal identity thresholds to discriminate filamentous fungal species were predicted as 99.6 % for ITS and 99.8 % for LSU. We showed that 17 % and 18 % of the species could not be discriminated by the ITS and LSU genetic markers, respectively. Among them, ∼8 % were indistinguishable using both genetic markers. ITS has been shown to outperform LSU in filamentous fungal species discrimination with a probability of correct identification of 82 % vs. 77.6 %, and a clustering quality value of 84 % vs. 77.7 %. At higher taxonomic classifications, LSU has been shown to have a better discriminatory power than ITS. With a clustering quality value of 80 %, LSU outperformed ITS in identifying filamentous fungi at the ordinal level. At the generic level, the clustering quality values produced by both genetic markers were low, indicating the necessity for taxonomic revisions at genus level and, likely, for applying more conserved genetic markers or even whole genomes. The taxonomic thresholds predicted for filamentous fungal identification at the genus, family, order and class levels were 94.3 %, 88.5 %, 81.2 % and 80.9 % based on ITS barcodes, and 98.2 %, 96.2 %, 94.7 % and 92.7 % based on LSU barcodes. The DNA barcodes used in this study have been deposited to GenBank and will also be publicly available at the Westerdijk Institute's website as reference sequences for fungal identification, marking an unprecedented data release event in global fungal barcoding efforts to date.
Keywords: Automated curation; Biological resource centre; Fungi; ITS; LSU; Taxonomic thresholds.
Figures

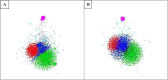

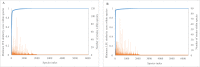

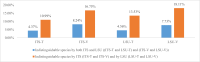

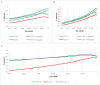



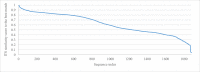
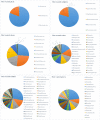
References
-
- Blaalid R., Kumar S., Nilsson R.H. ITS1 versus ITS2 as DNA metabarcodes for fungi. Molecular Ecology Resources. 2013;13:218–224. - PubMed
-
- Boon E., Zimmerman E., Lang B.F. Intra-isolate genome variation in arbuscular mycorrhizal fungi persists in the transcriptome. Journal of Evolutionary Biology. 2010;23:1519–1527. - PubMed
-
- Botschuijver S., Roeselers G., Levin E. Intestinal Fungal Dysbiosis Associates With Visceral Hypersensitivity in Patients With Irritable Bowel Syndrome and Rats. Gastroenterology. 2017;153:1026–1039. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
