Evolution on the Biophysical Fitness Landscape of an RNA Virus
- PMID: 29955873
- PMCID: PMC6188569
- DOI: 10.1093/molbev/msy131
Evolution on the Biophysical Fitness Landscape of an RNA Virus
Abstract
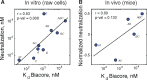
Viral evolutionary pathways are determined by the fitness landscape, which maps viral genotype to fitness. However, a quantitative description of the landscape and the evolutionary forces on it remain elusive. Here, we apply a biophysical fitness model based on capsid folding stability and antibody binding affinity to predict the evolutionary pathway of norovirus escaping a neutralizing antibody. The model is validated by experimental evolution in bulk culture and in a drop-based microfluidics that propagates millions of independent small viral subpopulations. We demonstrate that along the axis of binding affinity, selection for escape variants and drift due to random mutations have the same direction, an atypical case in evolution. However, along folding stability, selection and drift are opposing forces whose balance is tuned by viral population size. Our results demonstrate that predictable epistatic tradeoffs between molecular traits of viral proteins shape viral evolution.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

