A novel scoring system for acute myeloid leukemia risk assessment based on the expression levels of six genes
- PMID: 29956722
- PMCID: PMC6089755
- DOI: 10.3892/ijmm.2018.3739
A novel scoring system for acute myeloid leukemia risk assessment based on the expression levels of six genes
Abstract
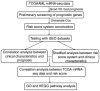
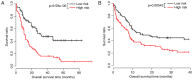
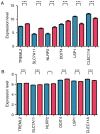
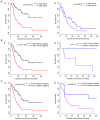
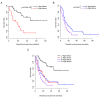
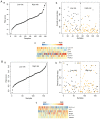
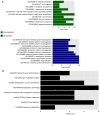
Acute myeloid leukemia (AML) is the most common type of acute leukemia and is a heterogeneous clonal disorder. At present, the pathogenesis of AML and potential methods to effectively prevent AML have become areas of interest in research. In the present study, two messenger ribonucleic acid sequencing datasets of patients with AML were downloaded from the Cancer Genome Atlas and Gene Expression Omnibus databases. The differentially expressed genes (DEGs) of the poor and good prognosis groups were screened using the Linear Models for Microarray Data package, and the prognosis‑related genes were screened using univariate Cox regression analysis. A total of 206 significant DEGs were identified. Following univariate and multivariate Cox regression analysis, 14 genes significantly associated with prognosis were screened and six of these genes, including triggering receptor expressed on myeloid cells 2 (TREML2), cysteine‑glutamate transporter (SLC7A11), NACHT, LRR, and PYD domains‑containing protein 2 (NLRP2), DNA damage‑inducible transcript 4 protein (DDIT4), lymphocyte‑specific protein 1 (LSP1) and C‑type lectin domain family 11 member A (CLEC11A), were used to construct model equations for risk assessment. The prognostic scoring system was used to evaluate risk for each patient, and the results showed that patients in the low‑risk group had a longer survival time, compared with those in the high‑risk group (P=9.59e‑06 for the training dataset and P=0.00543 for the validation dataset). A total of eight main Kyoto Encyclopedia of Genes and Genomes pathways were identified, the top three of which were hematopoietic cell lineage, focal adhesion, and regulation of actin cytoskeleton. Taken together, the results showed that the scoring system established in the present study was credible and that the six genes were identified, which were significantly associated with the risk assessment of AML, offer potential as prognostic biomarkers. These findings may provide clues for further clarifying the pathogenesis of AML.
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

