lncRNA-ATB functions as a competing endogenous RNA to promote YAP1 by sponging miR-590-5p in malignant melanoma
- PMID: 29956757
- PMCID: PMC6065447
- DOI: 10.3892/ijo.2018.4454
lncRNA-ATB functions as a competing endogenous RNA to promote YAP1 by sponging miR-590-5p in malignant melanoma
Abstract
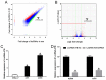
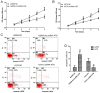
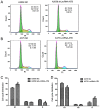
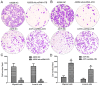
The critical long non‑coding RNAs (lncRNAs) involved in the carcinogenesis and progression of malignant melanoma (MM) have not been fully investigated. In the present study, it was identified that lncRNA activated by transforming growth factor‑β (lncRNA‑ATB) was upregulated in MM tissues and cells compared with benign nevus cells and human melanocytes, via comparative lncRNA screening from Gene Expression Omnibus datasets and reverse transcription‑quantitative polymerase chain reaction analysis. Furthermore, lncRNA‑ATB promoted the cell proliferation, cell migration, and cell invasion of MM cells in vitro, and tumor growth in vivo. It was additionally identified that lncRNA‑ATB attenuated cell cycle arrest and inhibited cellular apoptosis in MM cells. Finally, it was demonstrated that lncRNA‑ATB functions as a competing endogenous RNA (ceRNA) to enhance Yes associated protein 1 expression by competitively sponging microRNA miR‑590‑5p in MM cells. In conclusion, the present study revealed the expression and roles of lncRNA‑ATB in MM, and indicated that lncRNA‑ATB functions as a ceRNA to promote MM proliferation and invasion by sponging miR‑590‑5p.
Figures


Similar articles
-
miR‑590‑5p inhibits tumor growth in malignant melanoma by suppressing YAP1 expression.Oncol Rep. 2018 Oct;40(4):2056-2066. doi: 10.3892/or.2018.6633. Epub 2018 Aug 7. Oncol Rep. 2018. PMID: 30106445 Free PMC article.
-
Long non-coding RNA XIST promotes osteosarcoma progression by targeting YAP via miR-195-5p.J Cell Biochem. 2018 Jul;119(7):5646-5656. doi: 10.1002/jcb.26743. Epub 2018 Mar 25. J Cell Biochem. 2018. Retraction in: J Cell Biochem. 2021 Apr;122(3-4):485. doi: 10.1002/jcb.29872. PMID: 29384226 Retracted.
-
Long noncoding RNA kcna3 inhibits the progression of colorectal carcinoma through down-regulating YAP1 expression.Biomed Pharmacother. 2018 Nov;107:382-389. doi: 10.1016/j.biopha.2018.07.118. Epub 2018 Aug 9. Biomed Pharmacother. 2018. PMID: 30099342
-
The role of microRNAs and long non-coding RNAs in the pathology, diagnosis, and management of melanoma.Arch Biochem Biophys. 2014 Dec 1;563:60-70. doi: 10.1016/j.abb.2014.07.022. Epub 2014 Jul 24. Arch Biochem Biophys. 2014. PMID: 25065585 Free PMC article. Review.
-
Unravelling the Regulatory Roles of lncRNAs in Melanoma: From Mechanistic Insights to Target Selection.Int J Mol Sci. 2025 Feb 27;26(5):2126. doi: 10.3390/ijms26052126. Int J Mol Sci. 2025. PMID: 40076754 Free PMC article. Review.
Cited by
-
Long non-coding RNA KRT16P2/miR-1294/EGFR axis regulates laryngeal squamous cell carcinoma cell aggressiveness.Am J Transl Res. 2020 Jun 15;12(6):2939-2955. eCollection 2020. Am J Transl Res. 2020. PMID: 32655821 Free PMC article.
-
Expression and predictive value of miR-489 and miR-21 in melanoma metastasis.World J Clin Cases. 2019 Oct 6;7(19):2930-2941. doi: 10.12998/wjcc.v7.i19.2930. World J Clin Cases. 2019. PMID: 31624741 Free PMC article.
-
LncRNA TTN-AS1 promotes the progression of oral squamous cell carcinoma via miR-411-3p/NFAT5 axis.Cancer Cell Int. 2020 Aug 28;20:415. doi: 10.1186/s12935-020-01378-6. eCollection 2020. Cancer Cell Int. 2020. PMID: 32863773 Free PMC article.
-
Long noncoding RNA FAM83A-AS1 facilitates hepatocellular carcinoma progression by binding with NOP58 to enhance the mRNA stability of FAM83A.Biosci Rep. 2019 Nov 29;39(11):BSR20192550. doi: 10.1042/BSR20192550. Biosci Rep. 2019. PMID: 31696213 Free PMC article.
-
Linc00961 inhibits the proliferation and invasion of skin melanoma by targeting the miR‑367/PTEN axis.Int J Oncol. 2019 Sep;55(3):708-720. doi: 10.3892/ijo.2019.4848. Epub 2019 Jul 25. Int J Oncol. 2019. PMID: 31364744 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

